| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,417,224 – 17,417,325 |
| Length | 101 |
| Max. P | 0.551835 |

| Location | 17,417,224 – 17,417,325 |
|---|---|
| Length | 101 |
| Sequences | 13 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 67.99 |
| Shannon entropy | 0.72578 |
| G+C content | 0.32784 |
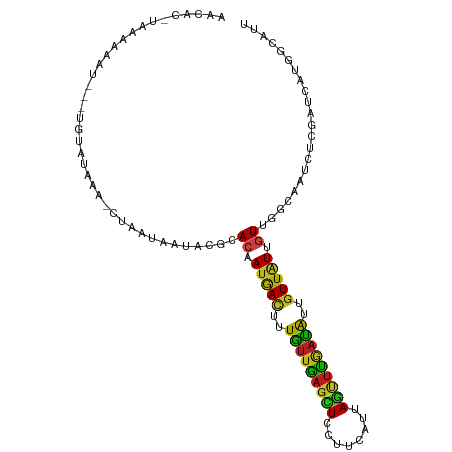
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -8.76 |
| Energy contribution | -7.61 |
| Covariance contribution | -1.15 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.36 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17417224 101 + 24543557 AACAG-UAUAAAAU---UGAGUACA-CUAAGAAUACGCACUAUGACUUUGUUGAGCUCCUUCAUAAGCUUGAUAUUGUUAUUGUUGGCAAUUUCGAUCAUGGCAUU ..(((-(.....))---)).((...-....(((...((((.(((((..(((..((((........))))..)))..))))).))..))...))).......))... ( -19.14, z-score = -0.27, R) >droAna3.scaffold_13337 8370114 100 + 23293914 ------UAAAGUAAGGAAAUAAUAUCUUUGCAGGACUUACAAUGACUUUGUUGAGUUCCUUCAUCAGUUUAAUAUUGUUAUUGUUGGCUAUCUCGAUCAUGGCAUU ------....(((((((.......))))))).......((((((((..((((((..(........)..))))))..))))))))..(((((.......)))))... ( -20.00, z-score = -0.81, R) >droEre2.scaffold_4784 9330726 101 - 25762168 AACAU-UAUUAAAC---UAUGUAUA-CUAAUAAUACGCACUAUGACUUUGUUGAGCUCCUUCAUUAGCUUGAUAUUGUUAUUGUUGGCAAUCUCGAUCAUGGCAUU ..(((-.(((....---...((((.-......))))((((.(((((..(((..((((........))))..)))..))))).))..))......))).)))..... ( -18.50, z-score = -1.04, R) >droYak2.chr3L 7548645 104 - 24197627 AGCAC-UAUAAAUUAUUUGGGUAUU-UUAAUAAUACGCACUAUGACUUUGUUGAGCUCCUUCAUUAGCUUGAUAUUGUUAUUGUUGGCAAUCUCGAUCAUGGCAUU .((..-..............(((((-.....)))))((((.(((((..(((..((((........))))..)))..))))).))..)).............))... ( -20.80, z-score = -0.88, R) >droSec1.super_0 9469663 101 + 21120651 AACAC-UCUAAAAU---UGGGUUAA-CUAAGAAUACGCACUAUGACUUUGUUGAGCUCCUUCAUUAGCUUGAUAUUGUUAUUGUUGGCAAUCUCGAUCAUGGCAUU .....-.(((..((---((((....-((((.(.........(((((..(((..((((........))))..)))..)))))).))))....))))))..))).... ( -18.50, z-score = -0.34, R) >droSim1.chr3L 16758165 101 + 22553184 AACAC-UAUAAAAU---UGGGUAAA-CUAAGAAUACGCACUAUGACUUUGUUGAGCUCCUUCAUUAGCUUGAUAUUGUUAUUGUUGGCAAUCUCGAUCAUGGCAUU ....(-(((....(---(((.....-))))......((((.(((((..(((..((((........))))..)))..))))).))..))..........)))).... ( -18.40, z-score = -0.33, R) >droPer1.super_35 503491 106 + 973955 GAUAAGUCAGAUAUAGAUAUACUUUCCUGCAGGUUCUUACGAUAACUUUGUUGAGUUCCUUCAUUAGUUUGAUGUUGUUGUUGUUGGCUAUUUCGAUCAUCGCAUU ((..((((((.....((((((((....((.(((..(((((((.....))).))))..))).))..)))...))))).......))))))...))............ ( -16.10, z-score = 1.26, R) >dp4.chrXR_group6 4094869 106 - 13314419 GAUAAGUCAGAUAUAGAUAUACUUUCCUGCAGGUUCUUACGAUAACUUUGUUGAGUUCCUUCAUUAGUUUGAUGUUGUUGUUGUUGGCUAUUUCGAUCAUCGCAUU ((..((((((.....((((((((....((.(((..(((((((.....))).))))..))).))..)))...))))).......))))))...))............ ( -16.10, z-score = 1.26, R) >droWil1.scaffold_180727 2490384 106 - 2741493 AUCUCAAAAUAUAUGGCUAAAUGCCUCUUUUACCACUUACAAUAACUUUAUUGAGCUCCUUCAUCAGUUUGAUAUUAUUAUUGUUGGCUAUCUCAAUCAUGGCAUU ..............(((.....))).............(((((((...(((..((((........))))..)))...)))))))..(((((.......)))))... ( -18.40, z-score = -1.80, R) >droVir3.scaffold_13049 19368982 100 - 25233164 -ACACGUAACCUGG----GUAUAUA-CUUUUCGCACAUACAAUAACUUUGUUGAGCUCUUUCAUUAAUUUGAUAUUGUUGUUGUUGGCAAUUUCGAUCAUGGCAUU -........(((((----((....)-))....((.((.((((((((..(((..((............))..)))..)))))))))))).........)).)).... ( -14.60, z-score = 0.70, R) >droMoj3.scaffold_6680 20568179 99 - 24764193 -ACGCAUAUCAAGA-----AGUAAC-AUUGUAAUACUCACAAUGACUUUAUUAAGUUCCUUCAUUAAUUUGAUAUUGUUGUUAUUGGCAAUUUCAAUCAUGGCAUU -..((.......((-----((...(-(((((.......))))))((((....))))..))))......((((.((((((......)))))).)))).....))... ( -17.50, z-score = -1.27, R) >droGri2.scaffold_15110 13215883 100 + 24565398 -----AUCAUAAGCAAGGUUCUAGA-UUUAUGGUACACACAAUAACUUUGUUGAGUUCUUUCAUCAAUUUGAUGUUGUUGUUGUUGGCAAUUUCAAUCAUAGCAUU -----.......((...((.(((..-....))).)).(((((((((..(((..((((........))))..)))..))))))).))...............))... ( -16.30, z-score = 0.38, R) >triCas2.ChLG2 6560647 98 - 12900155 --------AAUUUUAGUGGAAACAAUUGGAGAAAACCUACAACAAUAUUUUCCAAUUCUCGCAACAAAAGUUGGUUGUUUUUAUCAACGACCGAAAUCAGUGAAUU --------........((....))(((((((((..............)))))))))..((((........(((((((((......))))))))).....))))... ( -19.86, z-score = -1.58, R) >consensus AACAC_UAAAAAAU___UGUAUAAA_CUAAUAAUACGCACAAUGACUUUGUUGAGCUCCUUCAUUAGUUUGAUAUUGUUAUUGUUGGCAAUCUCGAUCAUGGCAUU ......................................((.(((((..(((((((((........)))))))))..))))).))...................... ( -8.76 = -7.61 + -1.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:37 2011