| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,402,779 – 17,402,869 |
| Length | 90 |
| Max. P | 0.948648 |

| Location | 17,402,779 – 17,402,869 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 62.15 |
| Shannon entropy | 0.72868 |
| G+C content | 0.59772 |
| Mean single sequence MFE | -20.31 |
| Consensus MFE | -10.27 |
| Energy contribution | -11.19 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17402779 90 - 24543557 CGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAAC--AACCA-------CAAUCACAA---UGACAACUCUAGCCGCAUAC ............((.(((((((.(((((...((((....)))))))))..))))..--.....-------...((....---.))....))).))....... ( -18.60, z-score = -1.74, R) >droSim1.chr3L 16743614 90 - 22553184 CGCCCACCUACCGCCAGAUACCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAAC--AACCA-------CAAUCACAA---UGACAACUCCAGCCGAUUGC .......................(((((...((((....)))))))))........--.....-------(((((....---((.......))...))))). ( -17.80, z-score = -1.19, R) >droSec1.super_0 9455397 90 - 21120651 CGCCCACCUACCGCCAGAUACCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAAC--AACCA-------CAAUCACAA---UGACAACUCCAGCCGCUUGC ............((..((.....(((((...((((....)))))))))..(((...--.....-------..)))....---.......))..))....... ( -15.90, z-score = -0.10, R) >droYak2.chr3L 7531852 96 + 24197627 CGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGCCAUGAUAAC--AACCA-CAACCGCAAUCACAA---UGACAACUCCAGCCGCUUGC ................(.((((.(((((...((((....)))))))))..)))).)--.....-.....((((.(....---((.......))...).)))) ( -17.40, z-score = 0.07, R) >droAna3.scaffold_13337 8356480 102 - 23293914 CGCCCACUUAUAGGCAGAUAUCCGGCGCCAUGGCAACCCUGCCGCGCCAUGAAAACCACACCAACAAUAAUAAUCACAAAAGCGGCGAUUCCGCUCGUCUGC .............((((((....(((((...((((....)))))))))................................(((((.....))))).)))))) ( -28.30, z-score = -1.85, R) >droWil1.scaffold_180698 1652626 87 + 11422946 CGCCUAUCUAUCGUCAAAUCUCCGGGGCCAUGGCCACCUUACCGCGAAAUGAACCACGCGACACUAG--UGGAGCGGAUCAACGGUUGC------------- ((((((.(((..(((........(((((....))).)).....(((..........))))))..)))--))).))).............------------- ( -22.10, z-score = 0.56, R) >droMoj3.scaffold_6680 20550749 69 + 24764193 CGCCCAUCUACAAACACAUAUCCGGCGGCAUGGCCACCUUGCCACGCCCCGACAGC--GAGCA------UCGCCUGC------------------------- ......................(((.(((.((((......)))).)))))).((((--(....------.)).))).------------------------- ( -20.00, z-score = -1.34, R) >droGri2.scaffold_15110 13203245 69 - 24565398 CGCUCACCUAUAAACAAAUAUCCGGCGGCAUGGCGACACUGCCACGCCCCGACGCG--GAGCA------GCGACUAC------------------------- .((((..((((......)))..(((.(((.(((((....))))).))))))..)..--)))).------........------------------------- ( -22.40, z-score = -2.12, R) >consensus CGCCCACCUACCGCCAGAUAUCCGGCGCCAUGGCAACUCUGCCGCGCCACGAUAAC__AACCA_______CAAUCACAA___UGACAACUCCAGCCG__UGC .......................(((((...((((....)))))))))...................................................... (-10.27 = -11.19 + 0.92)

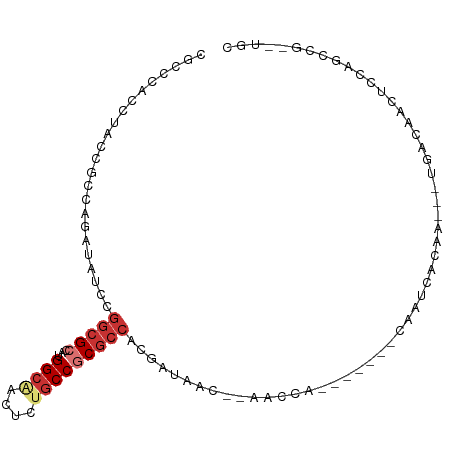
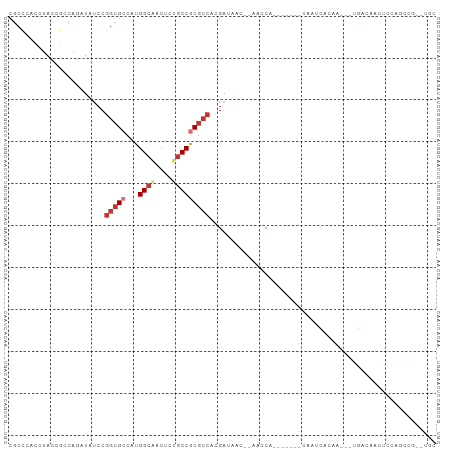
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:36 2011