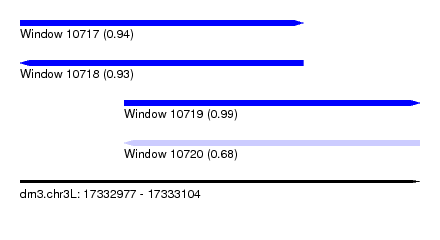
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,332,977 – 17,333,104 |
| Length | 127 |
| Max. P | 0.990371 |

| Location | 17,332,977 – 17,333,067 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 72.92 |
| Shannon entropy | 0.55133 |
| G+C content | 0.52180 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
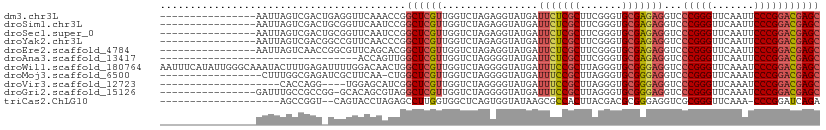
>dm3.chr3L 17332977 90 + 24543557 ---UUAUACAUUCCUUAGCGCACUGGGAUGACUGGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ---.....((((((..........))))))...((((...((((.((........)).)))).((((((.......))))))......)))). ( -29.10, z-score = -1.29, R) >droSim1.chr3L 16671138 80 + 22553184 ----UUUAAAUUCUUUAGCGCACUUGG---------GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ----............(((.(....).---------))).((((.((........)).)))).((((((.......))))))........... ( -21.50, z-score = -0.63, R) >droSec1.super_0 9384313 81 + 21120651 ---UUUUACAUUCUUCAGCGCACUUGG---------GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ---.............(((.(....).---------))).((((.((........)).)))).((((((.......))))))........... ( -21.60, z-score = -0.66, R) >droYak2.chr3L 7461076 78 - 24197627 ------AAUUAUUCUCAUUCCACUUGG---------GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ------...................((---------....((((.((........)).)))).((((((.......))))))......))... ( -20.00, z-score = -0.54, R) >droEre2.scaffold_4784 9246112 78 - 25762168 ------AAAUGUUCUUUGUCCACUUGG---------GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ------...........((((....))---------))..((((.((........)).)))).((((((.......))))))........... ( -23.00, z-score = -0.99, R) >droAna3.scaffold_13417 1014316 91 + 6960332 --UGGAAUAAAAUAUAAUAAACAAAAAUAAUUCAGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCU --...............................((((...((((.((........)).)))).((((((.......)))))).......)))) ( -22.90, z-score = -1.41, R) >droWil1.scaffold_180764 656387 73 - 3949147 -----------GAAUAAUCAUAUUUGG---------GCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCU -----------..............((---------(...((((.((........)).))))...((((.......)))).........))). ( -19.60, z-score = -0.34, R) >droVir3.scaffold_12723 3312776 84 - 5802038 AUUUGGUAAAAAGCUUAUUUUCCUUGG---------GCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCU ....((((...((((((.......)))---------))).((((.((........)).))))...((((.......))))......))))... ( -23.10, z-score = -0.26, R) >anoGam1.chr2R 11394264 86 + 62725911 -----ACGUAAGUACAACAAA--AAAGAAAUCGCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCU -----((....))........--...........(((.....(((((.......)))))((..((((((.......)))))).))....))). ( -23.70, z-score = -1.97, R) >consensus ______UAAAAUUCUUAGCACACUUGG_________GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCU ..........................................(((((.......)))))((..((((((.......)))))).))........ (-17.47 = -17.34 + -0.12)
| Location | 17,332,977 – 17,333,067 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 72.92 |
| Shannon entropy | 0.55133 |
| G+C content | 0.52180 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -20.55 |
| Energy contribution | -19.78 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17332977 90 - 24543557 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCCAGUCAUCCCAGUGCGCUAAGGAAUGUAUAA--- ...((.(((((((((((.......))))))(((.(((((.......)))))....)))...))))).)).(((((........)))))..--- ( -27.70, z-score = -0.08, R) >droSim1.chr3L 16671138 80 - 22553184 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC---------CCAAGUGCGCUAAAGAAUUUAAA---- .((((......))))..(((.((((((...(((.(((((.......)))))....))---------)....))))))...)))......---- ( -24.10, z-score = -0.31, R) >droSec1.super_0 9384313 81 - 21120651 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC---------CCAAGUGCGCUGAAGAAUGUAAAA--- .((((......)))).(((((((..(..(.(((.(((((.......)))))....))---------))..)..).)))))).........--- ( -27.10, z-score = -1.03, R) >droYak2.chr3L 7461076 78 + 24197627 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC---------CCAAGUGGAAUGAGAAUAAUU------ .......(.((((.(((((.((((.(....)((((.(((.......)))))))..))---------))))))))))).)........------ ( -25.60, z-score = -0.92, R) >droEre2.scaffold_4784 9246112 78 + 25762168 AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC---------CCAAGUGGACAAAGAACAUUU------ .(.(((....(((((((.......)))))))((((.(((.......)))))))..))---------))(((((.........)))))------ ( -24.30, z-score = -0.43, R) >droAna3.scaffold_13417 1014316 91 - 6960332 AGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCUGAAUUAUUUUUGUUUAUUAUAUUUUAUUCCA-- .(((((....(((((((.......)))))))((((.(((.......)))))))..)))))...............................-- ( -28.00, z-score = -1.26, R) >droWil1.scaffold_180764 656387 73 + 3949147 AGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGC---------CCAAAUAUGAUUAUUC----------- ..((((....(((((((.......)))))))((((.(((.......)))))))..))---------))..............----------- ( -26.30, z-score = -1.19, R) >droVir3.scaffold_12723 3312776 84 + 5802038 AGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGC---------CCAAGGAAAAUAAGCUUUUUACCAAAU ..((((....(((((((.......)))))))((((.(((.......)))))))..))---------))..((((((......)))).)).... ( -28.50, z-score = -0.82, R) >anoGam1.chr2R 11394264 86 - 62725911 AGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGCGAUUUCUUU--UUUGUUGUACUUACGU----- ..(((((..((((((((.......))))))((((.(((((((((.......))))))))).)))).....--...))..)))))....----- ( -30.70, z-score = -1.93, R) >consensus AGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC_________CCAAGUGCGAUAAGAAUUUUA______ ...((.....(((((((.......)))))))((((.(((.......))))))).............))......................... (-20.55 = -19.78 + -0.77)

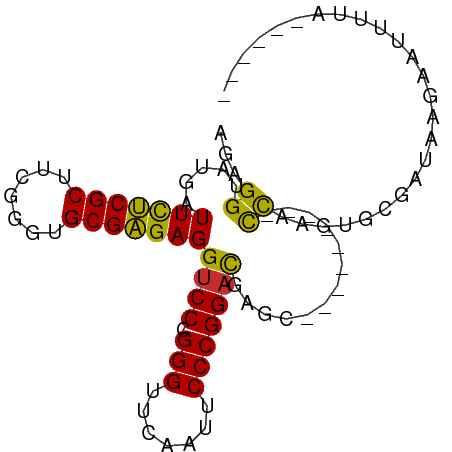
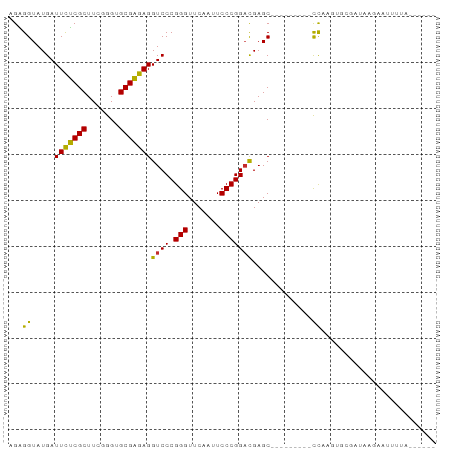
| Location | 17,333,010 – 17,333,104 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Shannon entropy | 0.52220 |
| G+C content | 0.57840 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -21.28 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17333010 94 + 24543557 GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGGUUUGAACCUCAGUCGACUAAUU---------------- (((((((((((.......)))))((..((((((.......)))))).))..............)))))).(((....)))..............---------------- ( -28.40, z-score = -1.57, R) >droSim1.chr3L 16671161 94 + 22553184 GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGGAUUGAACCGCAGUCGACUAAUU---------------- (((((((((((.......)))))((..((((((.......)))))).))..............))))))..(((((.....)))))........---------------- ( -30.60, z-score = -2.28, R) >droSec1.super_0 9384337 94 + 21120651 GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGGAUUGAACCGCAGUCGACUAAUU---------------- (((((((((((.......)))))((..((((((.......)))))).))..............))))))..(((((.....)))))........---------------- ( -30.60, z-score = -2.28, R) >droYak2.chr3L 7461097 94 - 24197627 GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGGGUUGAACGGCCGUCGACUAAUU---------------- (((((((((((.......)))))((..((((((.......)))))).))..............))))))..((((((.(....)))))))....---------------- ( -31.50, z-score = -1.28, R) >droEre2.scaffold_4784 9246133 94 - 25762168 GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGUGCUGAACGCCGGUUGACUAAUU---------------- (((((((((((.......)))))((..((((((.......)))))).))..............))))))((.((.....))))...........---------------- ( -29.40, z-score = -1.64, R) >droAna3.scaffold_13417 1014350 77 + 6960332 GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCAACUGGU--------------------------------- (((((((((((.......)))))((..((((((.......)))))).))..............))))))........--------------------------------- ( -26.70, z-score = -2.77, R) >droWil1.scaffold_180764 656403 110 - 3949147 GCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGUUGUCCAAAAUCUCAAAGUAUUUGCCCAAUAUGAAAUU (((((((((((.......)))))......((((.......))))...................))))))..((((..((((..(.....)..))))..))))........ ( -26.40, z-score = -1.63, R) >droMoj3.scaffold_6500 16677750 92 - 32352404 GCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAG-UUGAAGCGAUCUCGCCAAAG----------------- (((((((((((.......)))))......((((.......))))...................))))))...-.....(((....))).....----------------- ( -26.90, z-score = -1.84, R) >droVir3.scaffold_12723 3312803 86 - 5802038 GCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCGAUGCUCCA----CCUGGUG-------------------- (((((((((((.......)))))......((((.......))))...................)))))).......((.----...))..-------------------- ( -23.80, z-score = -0.82, R) >droGri2.scaffold_15126 7465660 93 + 8399593 GCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCUACGCUGUGC-CCGGCGGCAAAUC---------------- (((((((((((.......)))))......((((.......))))...................))))))...(((((...-.))))).......---------------- ( -30.10, z-score = -1.30, R) >triCas2.ChLG10 77815 87 - 8806720 UCUGAUCCGGG-UUUGAACCCGCGACCUCCCGCGUCGUAAGUGGCGCUUAUACCACUGAGCCACCAAGGCUCUAGGUACUG--ACCGGCU-------------------- .....((((((-(....))))).))....(((.(((((((((((........)))))(((((.....)))))....))).)--)))))..-------------------- ( -30.60, z-score = -1.42, R) >consensus GCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCGGUUUGAACCGC_GUCGACUAAUU________________ (((((((((((.......)))))......((((.......))))...................))))))......................................... (-21.28 = -20.90 + -0.38)
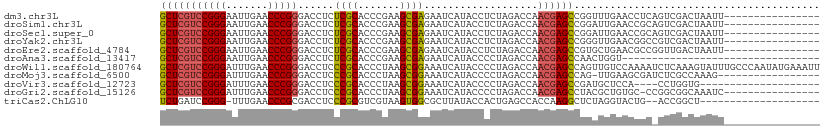
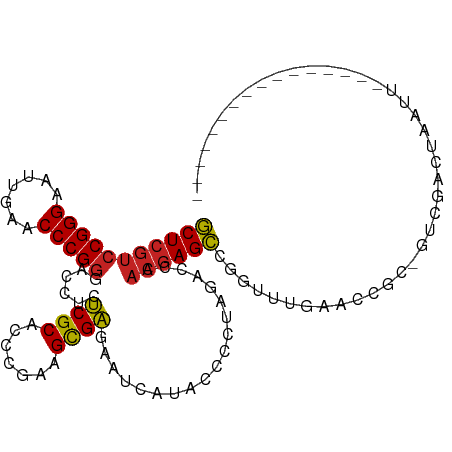
| Location | 17,333,010 – 17,333,104 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.46 |
| Shannon entropy | 0.52220 |
| G+C content | 0.57840 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -24.31 |
| Energy contribution | -24.26 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
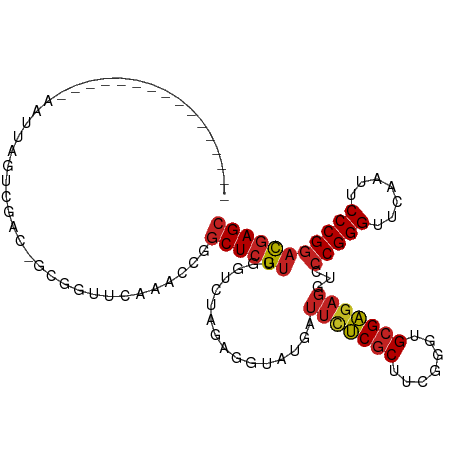
>dm3.chr3L 17333010 94 - 24543557 ----------------AAUUAGUCGACUGAGGUUCAAACCGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC ----------------..((((....))))(((....))).((((((..............(((((((((.......)))))).)))(((((.......))))))))))) ( -30.80, z-score = -0.05, R) >droSim1.chr3L 16671161 94 - 22553184 ----------------AAUUAGUCGACUGCGGUUCAAUCCGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC ----------------.............(((......)))((((((..............(((((((((.......)))))).)))(((((.......))))))))))) ( -30.70, z-score = 0.12, R) >droSec1.super_0 9384337 94 - 21120651 ----------------AAUUAGUCGACUGCGGUUCAAUCCGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC ----------------.............(((......)))((((((..............(((((((((.......)))))).)))(((((.......))))))))))) ( -30.70, z-score = 0.12, R) >droYak2.chr3L 7461097 94 + 24197627 ----------------AAUUAGUCGACGGCCGUUCAACCCGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC ----------------..((((((((((((((.......)))).)))))).))))........(((((((.......)))))))((((.(((.......))))))).... ( -36.20, z-score = -1.08, R) >droEre2.scaffold_4784 9246133 94 + 25762168 ----------------AAUUAGUCAACCGGCGUUCAGCACGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC ----------------...........((((.....)).))((((((..............(((((((((.......)))))).)))(((((.......))))))))))) ( -30.90, z-score = -0.09, R) >droAna3.scaffold_13417 1014350 77 - 6960332 ---------------------------------ACCAGUUGGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC ---------------------------------........(((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))) ( -29.10, z-score = -0.83, R) >droWil1.scaffold_180764 656403 110 + 3949147 AAUUUCAUAUUGGGCAAAUACUUUGAGAUUUUGGACAACUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGC ...........((((......((..(....)..))......))))(((.((((.(((((.(((..(((((.......))(((....)))))).))).))))))))).))) ( -31.90, z-score = 0.03, R) >droMoj3.scaffold_6500 16677750 92 + 32352404 -----------------CUUUGGCGAGAUCGCUUCAA-CUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGC -----------------....((((....))))....-...(((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))) ( -32.50, z-score = -0.20, R) >droVir3.scaffold_12723 3312803 86 + 5802038 --------------------CACCAGG----UGGAGCAUCGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGC --------------------.......----..((((....))))(((.((((.(((((.(((..(((((.......))(((....)))))).))).))))))))).))) ( -32.50, z-score = -0.74, R) >droGri2.scaffold_15126 7465660 93 - 8399593 ----------------GAUUUGCCGCCGG-GCACAGCGUAGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGC ----------------....((((.(..(-((.(((((......))))))))..).))))...(((((((.......)))))))((((.(((.......))))))).... ( -37.10, z-score = -0.69, R) >triCas2.ChLG10 77815 87 + 8806720 --------------------AGCCGGU--CAGUACCUAGAGCCUUGGUGGCUCAGUGGUAUAAGCGCCACUUACGACGCGGGAGGUCGCGGGUUCAAA-CCCGGAUCAGA --------------------..(((((--(.(((....(((((.....)))))((((((......)))))))))))).)))..((((.(((((....)-))))))))... ( -38.10, z-score = -2.57, R) >consensus ________________AAUUAGUCGAC_GCGGUUCAAACCGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGC .........................................((((((................(((((((.......)))))))...(((((.......))))))))))) (-24.31 = -24.26 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:31 2011