
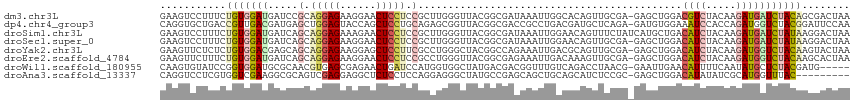
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,317,924 – 17,318,038 |
| Length | 114 |
| Max. P | 0.606921 |

| Location | 17,317,924 – 17,318,038 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.75 |
| Shannon entropy | 0.70312 |
| G+C content | 0.50816 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -10.30 |
| Energy contribution | -11.09 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.606921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17317924 114 - 24543557 GAAGUCCUUUCUGUGGAUGAUCCGCAGGAGAAGGAACUCCUCCGCUUGGGUUACGGCGAUAAAUUGGCACAGUUGCGA-GAGCUGGACGUCUACAAGAUGAUCUACAGCGACUAA ..((((....((((((((..((((.(((((......))))).)((((..((.((.((.(.....).))...)).))..-)))).)))((((.....)))))))))))).)))).. ( -35.40, z-score = -0.87, R) >dp4.chr4_group3 4463554 114 + 11692001 CAGGUGCUGACCGUUGACGAUGAGCUGGAGUACCAGCUCCUGCAGAGCGGUUACGGCGACCGCCUGACGAUGCUCAGA-GAUGUGGAAAUCCACCAGAUGGUCUACGGAUUCCAA ..((......((((.(((.(((((((((....)))))))(((..(((((((((.(((....)))))))..)))))...-...((((....))))))))).))).))))...)).. ( -43.60, z-score = -2.38, R) >droSim1.chr3L 16656006 115 - 22553184 GAAGUCCUUUCUGUGGAUGAUCAGCAGGAGAAAGAACUCCUCCGCUUGGGUUACGGCGAUAAAUUGGAACAGUUUCUAUCAUGCUGACAUCUACAAGAUGAUCUAUAAGGACUAA ..(((((((((((((((((.((((((((((......))))..((((........))))......(((((....)))))...))))))))))))).)))........))))))).. ( -37.60, z-score = -3.12, R) >droSec1.super_0 9369228 114 - 21120651 GAAGUCCUUUCUGUGGAUGAUCAGCAGGAGAAGGAACUCCUCCGCUUGGGUUACGGCGAUAAAUUGGAACAGUUGCGA-GAGCUGGACAUCUACAAGAUGAUCUAUAAGGACUAA ..(((((((((((((((((.(((((.((((......))))(((((....(((.(((.......))).)))....))).-))))))).))))))).)))........))))))).. ( -37.40, z-score = -2.45, R) >droYak2.chr3L 7445646 114 + 24197627 GAAGUUCUCUCUGUGGACGAGCAGCAGGAGAAGGAGCUCCUUCGCCUGGGCUACGGCCAGAAAUUGACGCAGUUGCGA-GAGCUGGACAUCUACAAGAUGGUCUACAAGUACUAA ..(((...((.(((((((.(((..((((.(((((....))))).))))(((....))).........(((....))).-..)))...((((.....))))))))))))).))).. ( -42.60, z-score = -2.02, R) >droEre2.scaffold_4784 9231205 114 + 25762168 GAAGUUCUUUCUGUGGAUGAUCAGCAGGAGAAGGAACUCCUCCGCCUGGGUUACGGCGAGAAAUUGACAAAGUUGCGA-GAGCUGGACAUCUACAAGAUGGUCUACAAGCACUAA ...((.((.((((((((((.(((((.((((......))))((((((........)))).)).................-..))))).))))))).))).))...))......... ( -33.60, z-score = -0.80, R) >droWil1.scaffold_180955 2099387 109 - 2875958 CAAGUGUAUCCGGUGGAUGCGCAACGUGAGCGAGAACUGAUCCAUGGUGGCUAUGACGACGGUUUGUCAGACCUAACG-GAAUUGAACAUUUUCAAUAUGCUCUACGAUG----- ...((((((((...))))))))..((((((((....(((((..((.((.(......).)).))..))))).((....)-).((((((....)))))).))))).)))...----- ( -33.40, z-score = -1.83, R) >droAna3.scaffold_13337 8273058 105 - 23293914 CAGGUCCUCGUGGUCGAAGGCGCAGUCGAGGAGGCUCUCCUCCAGGAGGGCUAUGCCGAGCAGCUGCAGCAUCUCCGC-GAGCUGGACAUAUAUCGCAUGGUUUAC--------- ...((((((((((..((..(((((((.(.((.((((((((....))))))))...))...).))))).)).)).))))-))...))))..................--------- ( -40.70, z-score = -0.59, R) >consensus GAAGUCCUUUCUGUGGAUGAUCAGCAGGAGAAGGAACUCCUCCGCUUGGGUUACGGCGAGAAAUUGACACAGUUGCGA_GAGCUGGACAUCUACAAGAUGGUCUACAAGGACUAA ...........(((((((.....(.(((((......))))).)............................................((((.....)))))))))))........ (-10.30 = -11.09 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:26 2011