| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,293,310 – 17,293,443 |
| Length | 133 |
| Max. P | 0.989312 |

| Location | 17,293,310 – 17,293,415 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Shannon entropy | 0.38560 |
| G+C content | 0.44282 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.892145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
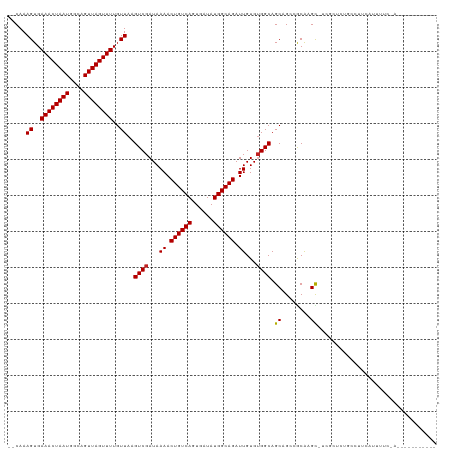
>dm3.chr3L 17293310 105 + 24543557 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUGCCUUCAUUUUCUA----------- --.((((((((.....((((((((....(((......)))...)))))))).......((((((.((((((((....))))))...-.)).))))))))))))))...----------- ( -33.00, z-score = -2.21, R) >droSim1.chr3L 16629986 105 + 22553184 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUGCCUUCAUUUUCUA----------- --.((((((((.....((((((((....(((......)))...)))))))).......((((((.((((((((....))))))...-.)).))))))))))))))...----------- ( -33.00, z-score = -2.21, R) >droSec1.super_0 9344391 105 + 21120651 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUGCCUUCAUUUUCUA----------- --.((((((((.....((((((((....(((......)))...)))))))).......((((((.((((((((....))))))...-.)).))))))))))))))...----------- ( -33.00, z-score = -2.21, R) >droYak2.chr3L 7419473 105 - 24197627 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUGCCUUCAUUUUCUA----------- --.((((((((.....((((((((....(((......)))...)))))))).......((((((.((((((((....))))))...-.)).))))))))))))))...----------- ( -33.00, z-score = -2.21, R) >droEre2.scaffold_4784 9205789 105 - 25762168 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUGCCUUCAUUUUCCA----------- --.((((((((.....((((((((....(((......)))...)))))))).......((((((.((((((((....))))))...-.)).))))))))))))))...----------- ( -33.00, z-score = -2.20, R) >droAna3.scaffold_13337 8251513 102 + 23293914 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCCUCCCAUCCUUCGCUG-------------- --...((..((((((((....))))))))..)).....((((....))))(((((.((((.((....((((((....))))))...-..)).))))...))))).-------------- ( -29.30, z-score = -1.77, R) >dp4.chrXR_group6 3961936 117 - 13314419 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCACAGGCCUCACUCCGUUUCUUUUUCCCUGCCGCAAAAGC --...............(((((.((((((((((.....))))))))))..(((((...(((......((((((....)))))).))))))))..............)))))........ ( -32.00, z-score = -1.06, R) >droPer1.super_35 373186 117 + 973955 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCACAGGCCUCACUCCGUUUCUUUUUCCCUGCCGCAAAAGC --...............(((((.((((((((((.....))))))))))..(((((...(((......((((((....)))))).))))))))..............)))))........ ( -32.00, z-score = -1.06, R) >droWil1.scaffold_180955 2071416 109 + 2875958 AGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGAGGCAGCAGCCGCAAGUCCCCACCGCCCCCUCACCUCCUC---------- .....((..((((((((....))))))))..))............((((((......))))))...((((((..((.(.(....)......).))..))))))......---------- ( -25.90, z-score = -0.65, R) >droMoj3.scaffold_6680 20431674 84 - 24764193 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCUG--GCGGGUCC------------------------------- --...((..((((((((....))))))))..))............((((((((....(.(((.....))).)..))))--))))....------------------------------- ( -20.80, z-score = -1.08, R) >droVir3.scaffold_13049 19226336 86 - 25233164 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCUG--GCGGGUAUCC----------------------------- --...((..((((((((....))))))))..)).....((((...((((((((....(.(((.....))).)..))))--)))).)))).----------------------------- ( -23.70, z-score = -2.12, R) >droGri2.scaffold_15110 13110085 84 + 24565398 --CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCUG--ACGGGUCG------------------------------- --...((..((((((((....))))))))..))....((((....((((((((....(.(((.....))).)..))))--))))))))------------------------------- ( -21.80, z-score = -1.64, R) >consensus __CAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC_UCGCUCUGCCUUCAUUUUC_A___________ .....((..((((((((....))))))))..))..((((...((.((((((......)))))).))...)))).............................................. (-20.09 = -20.18 + 0.08)


| Location | 17,293,348 – 17,293,443 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 65.89 |
| Shannon entropy | 0.61347 |
| G+C content | 0.46508 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -16.39 |
| Energy contribution | -16.36 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
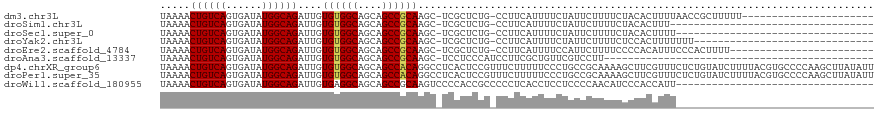
>dm3.chr3L 17293348 95 + 24543557 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUG-CCUUCAUUUUCUAUUCUUUUCUACACUUUUAACCGCUUUUU---------------------- .......(..(((((...((((((.((((((((....))))))...-.)).))))-)).)))))..)..............................---------------------- ( -22.10, z-score = -1.18, R) >droSim1.chr3L 16630024 83 + 22553184 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUG-CCUUCAUUUUCUAUUCUUUUCUACACUUU---------------------------------- .......(..(((((...((((((.((((((((....))))))...-.)).))))-)).)))))..)..................---------------------------------- ( -22.10, z-score = -1.62, R) >droSec1.super_0 9344429 84 + 21120651 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUG-CCUUCAUUUUCUAUUCUUUUCUACACUUUU--------------------------------- .......(..(((((...((((((.((((((((....))))))...-.)).))))-)).)))))..)...................--------------------------------- ( -22.10, z-score = -1.65, R) >droYak2.chr3L 7419511 87 - 24197627 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUG-CCUUCAUUUUCUAUUCUUUUCUCCACUUUUUUU------------------------------ .......(..(((((...((((((.((((((((....))))))...-.)).))))-)).)))))..)......................------------------------------ ( -22.10, z-score = -1.78, R) >droEre2.scaffold_4784 9205827 93 - 25762168 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCGCUCUG-CCUUCAUUUUCCAUUCUUUUCCCCACAUUUCCCACUUUU------------------------ ..........(((((...((((((.((((((((....))))))...-.)).))))-)).)))))...............................------------------------ ( -21.90, z-score = -1.75, R) >droAna3.scaffold_13337 8251551 73 + 23293914 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC-UCCUCCCAUCCUUCGCUGUUCGUCCUU--------------------------------------------- ....((...((((((.((((.((....((((((....))))))...-..)).))))...))))))...))....--------------------------------------------- ( -21.50, z-score = -1.90, R) >dp4.chrXR_group6 3961974 119 - 13314419 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCACAGGCCUCACUCCGUUUCUUUUUCCCUGCCGCAAAAGCUUCGUUUCUCUGUAUCUUUUACGUGCCCCAAGCUUAUAUU .....((((((......)))))).(((((((((....))))).(((......................)))))))((((((........((((.....))))......))))))..... ( -25.29, z-score = -0.05, R) >droPer1.super_35 373224 119 + 973955 UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCACAGGCCUCACUCCGUUUCUUUUUCCCUGCCGCAAAAGCUUCGUUUCUCUGUAUCUUUUACGUGCCCCAAGCUUAUAUU .....((((((......)))))).(((((((((....))))).(((......................)))))))((((((........((((.....))))......))))))..... ( -25.29, z-score = -0.05, R) >droWil1.scaffold_180955 2071456 86 + 2875958 UAAAACUGUCAGUGAUAUGGCAGAUUGUGAGGCAGCAGCCGCAAGUCCCCACCGCCCCCUCACCUCCUCCCCAACAUCCCACCAUU--------------------------------- .....((((((......))))))...((((((..((.(.(....)......).))..)))))).......................--------------------------------- ( -17.40, z-score = -1.35, R) >consensus UAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGC_UCGCUCUG_CCUUCAUUUUCUAUUCUUUUCUCCACUUUU_________________________________ .....((((((......))))))....((((((....))))))............................................................................ (-16.39 = -16.36 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:22 2011