| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,262,846 – 17,262,940 |
| Length | 94 |
| Max. P | 0.970643 |

| Location | 17,262,846 – 17,262,940 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.68089 |
| G+C content | 0.60961 |
| Mean single sequence MFE | -34.23 |
| Consensus MFE | -18.47 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17262846 94 + 24543557 CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC-UCGUCCCU------UGG--AUUCACCACCAGGACUCCCACAC--UCAGCUCCUGUGC ....(((((....(((((((.....)))))))((.(((((.(((.(.-..((((..------(((--.....)))...))))...).)))--.)))))))))))) ( -36.80, z-score = -1.20, R) >droYak2.chr3L 7388310 96 - 24197627 CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC-UCGUCCCU------UGG--AUUCACCACCAGGACUCCCACACACUCAGCUCCUGUGC ....((((((...(((((((.....)))))))(.((..((((((.(.-..((((..------(((--.....)))...))))..)))))))..)).)..)))))) ( -39.00, z-score = -1.71, R) >droSec1.super_0 9314448 93 + 21120651 CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC-UCGUCCCU------UGG--AUUCACCAUCAGGACUCC-ACAC--UCAGCUCCAGUGC ....((((.....(((((((.....)))))))((.(((((.(((.(.-..((((..------(((--.....)))...))))..)-.)))--.))))))).)))) ( -37.10, z-score = -1.64, R) >droSim1.chr3L 16598834 93 + 22553184 CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC-UCGUCCCU------UGG--AUUCACCACCAGGACUCC-ACAC--UCAGCUCCAGUGC ....((((.....(((((((.....)))))))((.(((((.(((.(.-..((((..------(((--.....)))...))))..)-.)))--.))))))).)))) ( -36.40, z-score = -1.18, R) >droWil1.scaffold_180955 2032125 101 + 2875958 CAUCGCACAGUUGGCUGUUGUCCUGCGACAGCGGUGGUUGUGUGCGCAUUAUCCUACCUUUCUUC--AUCCAUCAUCAACAAGACAAUAC--UUGGAACCAGCCA ...((((((..(.(((((((((....))))))))).)...))))))...................--............((((......)--))).......... ( -23.80, z-score = 0.22, R) >droPer1.super_35 338810 89 + 973955 UAUCGCACAGUCGGCUGUCGUCCUGCGGCAGCGAAGGUUGUGUGCGC-UCGUCCU---------G--CCCCAUAGUCAGGAUUCAGGCCC----AGCUUCAUCUG ...((((((..(.(((((((.....)))))))...)..)))))).((-(.(.(((---------(--..((.......))...)))).).----)))........ ( -31.50, z-score = -1.13, R) >dp4.chrXR_group6 3927272 89 - 13314419 UAUCGCACAGUCGGCUGUCGUCCUGCGGCAGCGAAGGUUGUGUGCGC-UCGUCCU---------G--CCCCAUAGUCAGGAUUCAGGCCC----AGCUUCAUCUG ...((((((..(.(((((((.....)))))))...)..)))))).((-(.(.(((---------(--..((.......))...)))).).----)))........ ( -31.50, z-score = -1.13, R) >droAna3.scaffold_13337 8219101 87 + 23293914 CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC-UUGUCC---------------UGCCUGCCAGGACUCUGGCUC--UCAGCUCCUGCUA ...(((((((..((((((((.....)))))))..)..))))))).((-(.((((---------------((.....))))))...)))..--..(((....))). ( -34.50, z-score = -0.45, R) >droEre2.scaffold_4784 9176148 94 - 25762168 CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC-UCGUCCCU------UGG--ACUCACCACCAGGACUCCCACAC--UCAGCUCCUGUGC ....(((((....(((((((.....)))))))((.(((((.(((.(.-..((((..------(((--.....)))...))))...).)))--.)))))))))))) ( -36.80, z-score = -0.91, R) >droVir3.scaffold_13049 19193705 85 - 25233164 CAGCGCGCAGUUGGCUGCCGUCCUGCGACAGCGGUAGUUGUGUGCGC-UUGACUGGCCCCAGUCU---CCCUCUCCCAAC-CCCCAGCCA--------------- .((((((((..((((((((((.........)))))))))))))))))-).(((((....))))).---............-.........--------------- ( -31.00, z-score = -1.30, R) >droMoj3.scaffold_6680 20393047 94 - 24764193 CAGCGCGCAGUUGGCUGUCGUCCUGCGACCGCGUUAGUUGUGUGCGC-UUGACGGGCCUUUGUGG---CCCAAACCCA---CUCCAGCUAACUGAACUUUA---- ....(..((((((((((..(((..(((..((((.....))))..)))-..)))(((((.....))---))).......---...))))))))))..)....---- ( -40.40, z-score = -3.24, R) >droGri2.scaffold_15110 13081802 93 + 24565398 GAACGCGCAGUUGGCUGUCGUCCUGCGGCAGCGGUAGUUGUGUGCGC-UUGACUGGCCCCCAUCAGAACCAGUACCUAAAAGUGCCACUAGCCA----------- .(((.....))).(((((((.....)))))))(((....(((.((((-((.(((((..(......)..)))))......)))))))))..))).----------- ( -32.00, z-score = -0.40, R) >consensus CAUCGCACAGUUGGCUGUCGUCCUGCGGCAGCGGUGGUUGUGUGCGC_UCGUCCCU______UGG__ACCCACCACCAGGACUCCCACAC__UCAGCUCCAGUGC ...((((((..(.(((((((.....)))))))...)..))))))............................................................. (-18.47 = -17.93 + -0.55)

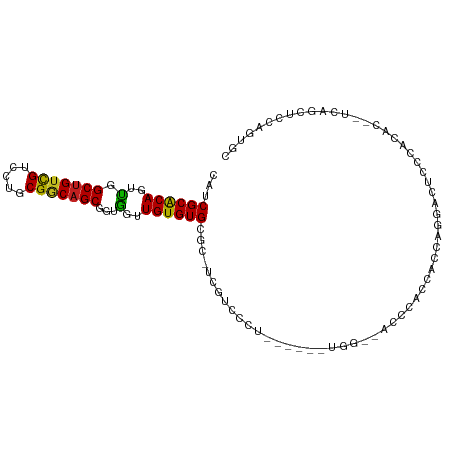
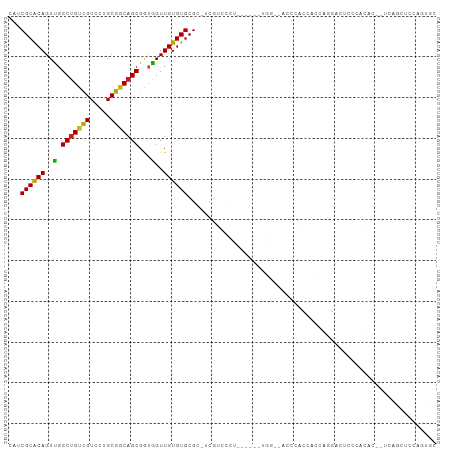
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:19 2011