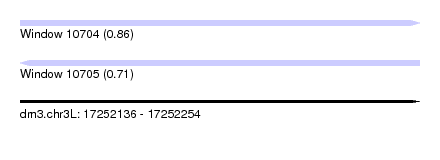
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,252,136 – 17,252,254 |
| Length | 118 |
| Max. P | 0.861537 |

| Location | 17,252,136 – 17,252,254 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Shannon entropy | 0.12678 |
| G+C content | 0.40217 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -22.40 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17252136 118 + 24543557 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCUACGAGG ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))..(--(((...)))) ( -26.20, z-score = -2.12, R) >droSim1.chr3L 16587436 118 + 22553184 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCUACGAGG ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))..(--(((...)))) ( -26.20, z-score = -2.12, R) >droSec1.super_0 9303154 118 + 21120651 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCUACGAGG ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))..(--(((...)))) ( -26.20, z-score = -2.12, R) >droYak2.chr3L 7377098 118 - 24197627 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCUACGAGG ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))..(--(((...)))) ( -26.20, z-score = -2.12, R) >droEre2.scaffold_4784 9165191 118 - 25762168 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCUACGAGG ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))..(--(((...)))) ( -26.20, z-score = -2.12, R) >droAna3.scaffold_13337 8208744 118 + 23293914 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCGAAGAGU ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))...--.......... ( -23.60, z-score = -1.34, R) >dp4.chrXR_group6 3915215 118 - 13314419 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCGUCGAGU .......(((((...((((.((((((((((((.((((....))))))))))............((........)).....)))))).)))).....((.....))...--...))))).. ( -24.20, z-score = -1.19, R) >droPer1.super_35 326652 118 + 973955 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC--CUCGUCGAGU .......(((((...((((.((((((((((((.((((....))))))))))............((........)).....)))))).)))).....((.....))...--...))))).. ( -24.20, z-score = -1.19, R) >droWil1.scaffold_180955 2015656 120 + 2875958 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUAUACGCCAUGGCCCUAUAAGG ...(((((.(....)((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))..))))).....((....))......... ( -24.20, z-score = -0.64, R) >droVir3.scaffold_13049 19181157 106 - 25233164 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUACGCC-------------- ...(((((.(....)((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))..)))))........-------------- ( -22.40, z-score = -1.77, R) >droMoj3.scaffold_6059 3242 107 - 6074 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUACGCCA------------- ...(((((.(....)((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))..))))).........------------- ( -22.40, z-score = -1.71, R) >droGri2.scaffold_15110 6655903 107 + 24565398 CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCA------------- ...(((...(((((.((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))...)))))...)))..------------- ( -23.60, z-score = -2.01, R) >consensus CACGCAAUCGGCAACGUGUCGCAUUUGUAGUUAGACAUAAUUGUUAAUUACCCGAAACCAUUACCACAUUAAAGGCCAUAAAAUGCAACACUAAUUGCUUUAUGCCAC__CUCUACGAGG ...(((((.(....)((((.((((((((((((.((((....))))))))))............((........)).....)))))).))))..)))))...................... (-22.40 = -22.40 + 0.00)


| Location | 17,252,136 – 17,252,254 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.12 |
| Shannon entropy | 0.12678 |
| G+C content | 0.40217 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17252136 118 - 24543557 CCUCGUAGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG ...(((((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..))..))))).. ( -32.90, z-score = -1.45, R) >droSim1.chr3L 16587436 118 - 22553184 CCUCGUAGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG ...(((((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..))..))))).. ( -32.90, z-score = -1.45, R) >droSec1.super_0 9303154 118 - 21120651 CCUCGUAGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG ...(((((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..))..))))).. ( -32.90, z-score = -1.45, R) >droYak2.chr3L 7377098 118 + 24197627 CCUCGUAGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG ...(((((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..))..))))).. ( -32.90, z-score = -1.45, R) >droEre2.scaffold_4784 9165191 118 + 25762168 CCUCGUAGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG ...(((((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..))..))))).. ( -32.90, z-score = -1.45, R) >droAna3.scaffold_13337 8208744 118 - 23293914 ACUCUUCGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG .(((...)))--...(((....(((((..(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).))))))))))))))))....)))... ( -30.10, z-score = -0.88, R) >dp4.chrXR_group6 3915215 118 + 13314419 ACUCGACGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG .....(((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..)).....))). ( -30.90, z-score = -0.80, R) >droPer1.super_35 326652 118 - 973955 ACUCGACGAG--GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG .....(((.(--(..((...((....)).(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).)))))))))))))..)).....))). ( -30.90, z-score = -0.80, R) >droWil1.scaffold_180955 2015656 120 - 2875958 CCUUAUAGGGCCAUGGCGUAUAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG (((....))).....(((((..(((((..(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).))))))))))))))))....))))). ( -31.40, z-score = -0.39, R) >droVir3.scaffold_13049 19181157 106 + 25233164 --------------GGCGUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG --------------.((((((.(((((..(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).))))))))))))))))...)))))). ( -31.10, z-score = -2.01, R) >droMoj3.scaffold_6059 3242 107 + 6074 -------------UGGCGUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG -------------..((((((.(((((..(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).))))))))))))))))...)))))). ( -31.10, z-score = -1.89, R) >droGri2.scaffold_15110 6655903 107 - 24565398 -------------UGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG -------------..(((....(((((..(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).))))))))))))))))....)))... ( -29.50, z-score = -1.56, R) >consensus CCUCGUAGAG__GUGGCAUAAAGCAAUUAGUGUUGCAUUUUAUGGCCUUUAAUGUGGUAAUGGUUUCGGGUAAUUAACAAUUAUGUCUAACUACAAAUGCGACACGUUGCCGAUUGCGUG ...............(((....(((((..(((((((((((....(((........)))..(((((..(.(((((.....))))).)..))))).))))))))))))))))....)))... (-29.51 = -29.33 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:18 2011