| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,518,812 – 5,518,877 |
| Length | 65 |
| Max. P | 0.921442 |

| Location | 5,518,812 – 5,518,877 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 71 |
| Reading direction | forward |
| Mean pairwise identity | 85.45 |
| Shannon entropy | 0.30475 |
| G+C content | 0.50117 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.921442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5518812 65 + 23011544 GCUCUACUGACACCAAUCGGGCGCCAUUUUGGAGAUGGACAACUUCUCGAUUGGCGAAUAACUUA------ ..........(.(((((((((..(((((.....))))).......))))))))).).........------ ( -16.80, z-score = -2.56, R) >droSim1.chr2L 5321493 65 + 22036055 GCUCUACUGACACCAAUCGGGCGCCAUUUUGGAGAUGGACAACUUCUCGAUUGGCGAAUAACUUA------ ..........(.(((((((((..(((((.....))))).......))))))))).).........------ ( -16.80, z-score = -2.56, R) >droSec1.super_5 3597315 65 + 5866729 GCUCUACUGACACCAAUCGGGCGCCAUUUUGGAGAUGGACAACUUCUCGAUUGGCGAAUAACUUA------ ..........(.(((((((((..(((((.....))))).......))))))))).).........------ ( -16.80, z-score = -2.56, R) >droYak2.chr2L 8648350 65 - 22324452 GCUCUACUGACACCAAUCGGGCGCCAUUUUGGAGAUGGACAACUUCUCGAUUGGCGAAUGACUCG------ ..........(.(((((((((..(((((.....))))).......))))))))).).........------ ( -16.80, z-score = -1.83, R) >droEre2.scaffold_4929 5604946 65 + 26641161 GCUCUACUGACACCAAUCGGGCGCCAUUUUGGAGAUGGACAACUUCUCCAUUGGCGAAUGACUCG------ ................(((..(((((...((((((.(.....).)))))).)))))..)))....------ ( -21.10, z-score = -3.65, R) >droAna3.scaffold_12916 6550382 65 - 16180835 GCUCUACUGACACCAAUCGGGCGCCAUCCUGGAAAUGGACAACUUUUCCAUUGGCGAAUGAUUCG------ ..............(((((..(((((...((((((.(.....).)))))).)))))..)))))..------ ( -19.30, z-score = -2.97, R) >dp4.chr4_group1 4836170 71 + 5278887 GCUCUACUGACACGAGUCGGGCGCCAUUCUGGAAAUGGACAACUUCUCGAUUGGCGAAUGAGGACUGCUAU .(((..(((((....))))).(((((.((.((((.........)))).)).)))))...)))......... ( -20.50, z-score = -1.89, R) >droPer1.super_5 6432775 71 - 6813705 GCUCUACUGACACGAGUCGGGCGCCAUUCUGGAAAUGGACAACUUCUCGAUUGGCGAAUGAGGACUGCUAU .(((..(((((....))))).(((((.((.((((.........)))).)).)))))...)))......... ( -20.50, z-score = -1.89, R) >droWil1.scaffold_180708 713416 65 + 12563649 ACUCUACUGACACGAAUCGGGCGCCAUUCUGGAAAUGGAUAACUUUUCGAUUGGCGAAUGAAUGA------ ................(((..(((((.((.(((((.(.....)))))))).)))))..)))....------ ( -15.60, z-score = -2.10, R) >droGri2.scaffold_15252 6010693 61 - 17193109 GCUCUACUGACACGAAUCGGGCGCCAUUCUGGAGAUGGACAACUUCUCCAUUGGCGAAUGA---------- ................(((..(((((...((((((.(.....).)))))).)))))..)))---------- ( -19.90, z-score = -2.98, R) >droVir3.scaffold_12963 487679 61 - 20206255 GCUCUACUGACACGAAUCGGGCGCCAUACUCGAAAUGGACGACUUCUCGAUAGGCGAAUGA---------- ................(((..((((.((.((((...(.....)...))))))))))..)))---------- ( -12.10, z-score = -0.19, R) >droMoj3.scaffold_6500 23486072 61 + 32352404 GCUCUACUGACACGAGUCGGGCGCCAUUCUAGAAAUGGACAAUUUCUCCAUAGGCGAAUGA---------- ......(((((....))))).((((.....((((((.....)))))).....)))).....---------- ( -16.20, z-score = -1.44, R) >consensus GCUCUACUGACACCAAUCGGGCGCCAUUCUGGAAAUGGACAACUUCUCGAUUGGCGAAUGACU_A______ ......((((......)))).(((((...(.((((.(.....).)))).).)))))............... (-12.10 = -12.36 + 0.26)
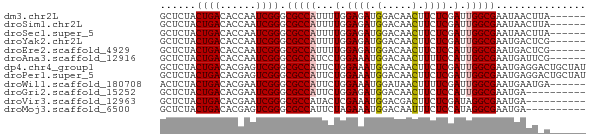
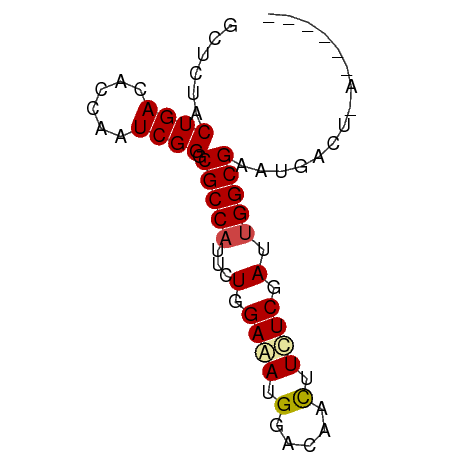
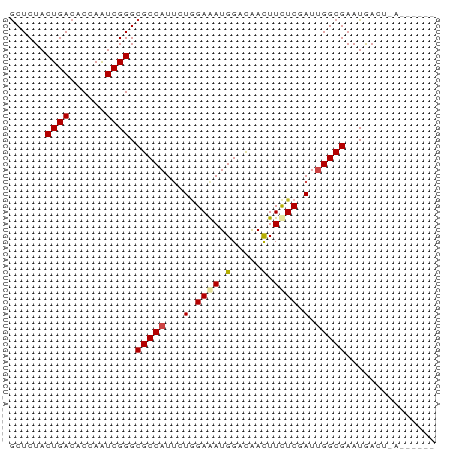
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:15 2011