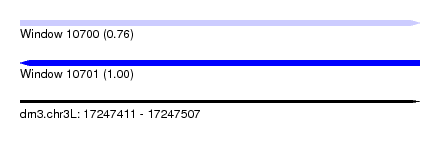
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,247,411 – 17,247,507 |
| Length | 96 |
| Max. P | 0.995617 |

| Location | 17,247,411 – 17,247,507 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 74.31 |
| Shannon entropy | 0.50580 |
| G+C content | 0.51711 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -16.92 |
| Energy contribution | -18.06 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.759044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
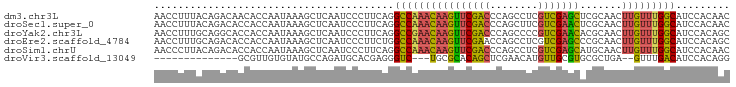
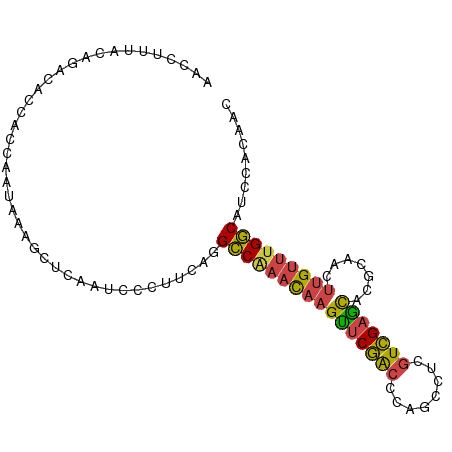
>dm3.chr3L 17247411 96 + 24543557 GUUGUGGAUGCCAAACAAGUUGCGAGCUCGACGAGGCUGGGUCGAACUUGUUUGGCCUGAAGGGAUUGAGCUUUAUUGGUGUUGUCUGUAAAGGUU .(((..(((((((((((((((.(((.((((.......))))))))))))))))))).....((.((..(......)..)).)))))..)))..... ( -29.70, z-score = -1.59, R) >droSec1.super_0 9298326 96 + 21120651 GUUGUGGAUGCCAAACAAGUUGCGAGUUCGACGAAGCUGGGUCGAACUUGUUUGGCCUGAAGGGAUUGAGCUUUAUUGGUGGUGUCUGUAAAGGUU .(((..((((((...(((...(((((((((((........))))))))))))))(((..((((.......))))...)))))))))..)))..... ( -32.60, z-score = -3.00, R) >droYak2.chr3L 7371785 96 - 24197627 GCUGUGGAUGCCAAACAAGUUGCGUGUUCGACGGGGCUGGGUCGAACUUGUUCGGCCUGAAGGGAUUGAGCUUUAUUGGUGGUGCCUGCAAAGGUU ((...((.((((((...((((.(((.....))).))))((((((((....))))))))((((........)))).))))))...)).))....... ( -28.20, z-score = 0.43, R) >droEre2.scaffold_4784 9160035 96 - 25762168 GCUGUGGAUGCCAAACAAGUUGCGGGCUCGACGAGGCUGGUUCGAACUUGUUUGGCCAGAAGGGAUUGAGCUUUAUUGGUGGUGUCUGCAAAGGUU ..((..(((((((((((((((.((((((.(......).))))))))))))))).(((((.((((......)))).)))))))))))..))...... ( -34.90, z-score = -2.18, R) >droSim1.chrU 12069401 96 + 15797150 GUUGUGGAUGCCAAACAAGUUGCAUGCUCGACGAGGCUGGGUCGAACUUGUUUGGCCUGAAGGGAUUGAGCUUUAUUGGUGGUGUCUGUAAGGGUU .(((..(((((((..(((.......((((((.......((((((((....)))))))).......))))))....))).)))))))..)))..... ( -29.64, z-score = -1.30, R) >droVir3.scaffold_13049 19175795 77 - 25233164 CCUGUGGAUGUCAAAC--UCAGCGCACGCAACAUGUUCGAGCUGUGCGCA---GACCCUCGUGCAUCUGGCAUACACAACGC-------------- ..((((.(((((....--...((((((((...........)).))))))(---((..(....)..)))))))).))))....-------------- ( -22.10, z-score = 0.20, R) >consensus GCUGUGGAUGCCAAACAAGUUGCGAGCUCGACGAGGCUGGGUCGAACUUGUUUGGCCUGAAGGGAUUGAGCUUUAUUGGUGGUGUCUGUAAAGGUU ..((..((((((((((((((.......(((((........))))))))))))))))...((((.......)))).........)))..))...... (-16.92 = -18.06 + 1.14)
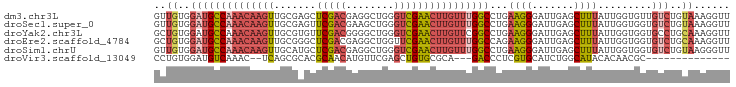
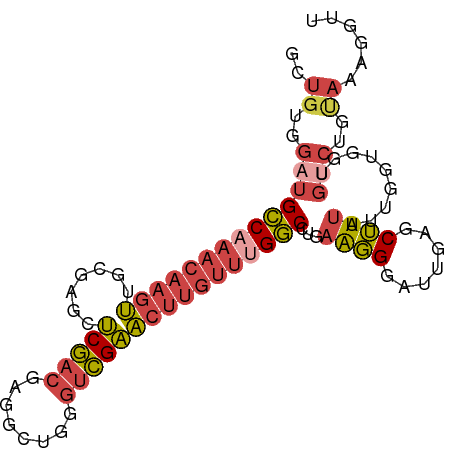
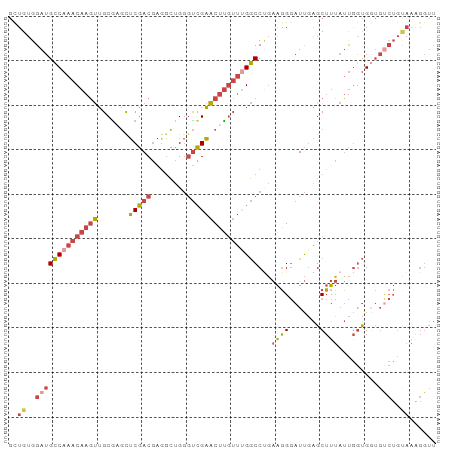
| Location | 17,247,411 – 17,247,507 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.31 |
| Shannon entropy | 0.50580 |
| G+C content | 0.51711 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -17.35 |
| Energy contribution | -17.08 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17247411 96 - 24543557 AACCUUUACAGACAACACCAAUAAAGCUCAAUCCCUUCAGGCCAAACAAGUUCGACCCAGCCUCGUCGAGCUCGCAACUUGUUUGGCAUCCACAAC ........................................((((((((((((((((........)))))(....).)))))))))))......... ( -21.00, z-score = -3.28, R) >droSec1.super_0 9298326 96 - 21120651 AACCUUUACAGACACCACCAAUAAAGCUCAAUCCCUUCAGGCCAAACAAGUUCGACCCAGCUUCGUCGAACUCGCAACUUGUUUGGCAUCCACAAC ........................................((((((((((((((((........))))).......)))))))))))......... ( -20.91, z-score = -3.71, R) >droYak2.chr3L 7371785 96 + 24197627 AACCUUUGCAGGCACCACCAAUAAAGCUCAAUCCCUUCAGGCCGAACAAGUUCGACCCAGCCCCGUCGAACACGCAACUUGUUUGGCAUCCACAGC ...(((((..((.....))..)))))..............((((((((((((((((........))))).......)))))))))))......... ( -23.21, z-score = -2.12, R) >droEre2.scaffold_4784 9160035 96 + 25762168 AACCUUUGCAGACACCACCAAUAAAGCUCAAUCCCUUCUGGCCAAACAAGUUCGAACCAGCCUCGUCGAGCCCGCAACUUGUUUGGCAUCCACAGC ........((((........................))))((((((((((((((.....((........)).)).))))))))))))......... ( -19.36, z-score = -1.15, R) >droSim1.chrU 12069401 96 - 15797150 AACCCUUACAGACACCACCAAUAAAGCUCAAUCCCUUCAGGCCAAACAAGUUCGACCCAGCCUCGUCGAGCAUGCAACUUGUUUGGCAUCCACAAC ........................................((((((((((((((((........)))))(....).)))))))))))......... ( -21.00, z-score = -3.15, R) >droVir3.scaffold_13049 19175795 77 + 25233164 --------------GCGUUGUGUAUGCCAGAUGCACGAGGGUC---UGCGCACAGCUCGAACAUGUUGCGUGCGCUGA--GUUUGACAUCCACAGG --------------.(.((((((((.....)))))))).).((---.((((((.((...........)))))))).))--................ ( -22.60, z-score = 1.27, R) >consensus AACCUUUACAGACACCACCAAUAAAGCUCAAUCCCUUCAGGCCAAACAAGUUCGACCCAGCCUCGUCGAGCACGCAACUUGUUUGGCAUCCACAAC ........................................((((((((((((((((........))))))).......)))))))))......... (-17.35 = -17.08 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:15 2011