
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,221,577 – 17,221,685 |
| Length | 108 |
| Max. P | 0.769184 |

| Location | 17,221,577 – 17,221,685 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.00 |
| Shannon entropy | 0.57319 |
| G+C content | 0.55148 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -14.02 |
| Energy contribution | -14.16 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.769184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
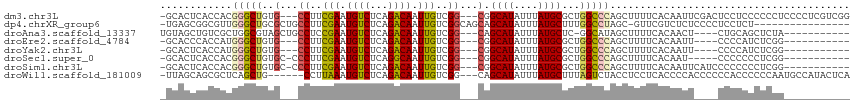
>dm3.chr3L 17221577 108 - 24543557 -GCACUCACCACGGGCUGUG---CCUUCGAAUGUCUCAGACAAUUGUCGG---CGGCAUAUUUAUGCGCUGGCCCAGCUUUUCACAAUUCGACUCCUCCCCCCUCCCCUCGUCGG -...........((((((((---((..(((.((((...)))).)))..))---).((((....)))))).)))))..............((((.................)))). ( -26.03, z-score = -0.31, R) >dp4.chrXR_group6 389181 97 + 13314419 -UGAGCGGCGUUGGGCUGCGCUGCCUUCGAAUGUCUCAGACAAUUGUCGGCAGCAGCAUAUUUAUGCUUUGGCCUAGC-GUUCGUCUCUCCCCUCCUCU---------------- -.(((((((((((((((..((((((..(((.((((...)))).)))..))))))(((((....)))))..))))))))-).))).)))...........---------------- ( -42.50, z-score = -4.25, R) >droAna3.scaffold_13337 7343580 96 - 23293914 UGUAGCUGUCGCUGGCGUAGCUGCCUCCGAAUGUCUCAGACAAUUGUCGG---CAGCAUAUUUAUGCUC-GGCAUAGCUUUUCACAACU----CUGCAGCUCUA----------- ((.((((((.(((((((((((((((..(((.((((...)))).)))..))---)))).....))))).)-)))))))))...)).....----...........----------- ( -33.90, z-score = -2.40, R) >droEre2.scaffold_4784 9134429 93 + 25762168 -GCACCCACCAUGGGCUGUG---CCUUCGAAUGUCUCAGACAAUUGUCGG---CGGCAUAUUUAUGCGCUGGCCCAGCUUUUCACAAUU----CCCCAUCUCGG----------- -....((...(((((.((((---((.((((.((((...))))....))))---.)))))).......((((...))))...........----.)))))...))----------- ( -24.20, z-score = -0.24, R) >droYak2.chr3L 7345263 93 + 24197627 -GCACUCACCAUGGGCUGUG---CCUUCGAAUGUCUCAGACAAUUGUCGG---CGGCAUAUUUAUGCGCUGGCCCAGCUUUUCACAAUU----CCCCAUCUCGG----------- -..........(((((((((---((..(((.((((...)))).)))..))---).((((....)))))).)))))).............----...........----------- ( -24.00, z-score = -0.38, R) >droSec1.super_0 9273649 94 - 21120651 -GCACUCACCACGGGCUGUGC-CCCUUCGAAUGUCUCAGGCAAUUGUCGG---CGGCAUAUUUAUGCGCUGGCCCAGCUUUUCACAAU-----CCCCCCCUCGG----------- -.......((..(((.(((((-(((..(((.(((.....))).)))..))---.)))))).......((((...))))..........-----....)))..))----------- ( -23.70, z-score = 0.68, R) >droSim1.chr3L 16556529 99 - 22553184 -GCACUCACCACGGGCUGUGC-CCCUUCGAAUGUCUCAGACAAUUGUCGG---CGGCAUAUUUAUGCGCUGGCCCAGCUUUUCACAAUUCAUCCCCCCCCUCGG----------- -.......((..(((.(((((-(((..(((.((((...)))).)))..))---.)))))).......((((...))))...................)))..))----------- ( -24.00, z-score = -0.18, R) >droWil1.scaffold_181009 428659 105 + 3585778 -UUAGCAGCGCUCAGCUG------CCUUAAAUGUCUCAGACAAUUGUCGG---CAGCAUAUUUAUGCUUUAGUCUACCUCCUCACCCCACCCCCCACCCCCCAAUGCCAUACUCA -...(((((.....))))------).............(((....)))((---((((((....))))....(....)...........................))))....... ( -18.30, z-score = -1.90, R) >consensus _GCACCCACCACGGGCUGUG___CCUUCGAAUGUCUCAGACAAUUGUCGG___CGGCAUAUUUAUGCGCUGGCCCAGCUUUUCACAAUU____CCCCCCCUCGG___________ ............(((((......((..(((.((((...)))).)))..)).....((((....))))...)))))........................................ (-14.02 = -14.16 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:11 2011