| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,219,157 – 17,219,251 |
| Length | 94 |
| Max. P | 0.931514 |

| Location | 17,219,157 – 17,219,251 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Shannon entropy | 0.42060 |
| G+C content | 0.40563 |
| Mean single sequence MFE | -22.89 |
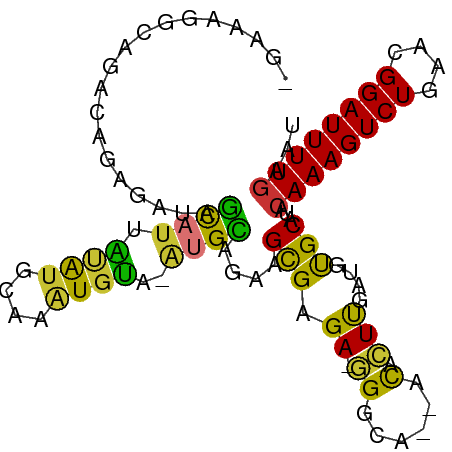
| Consensus MFE | -11.30 |
| Energy contribution | -10.18 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
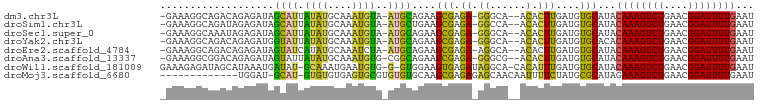
>dm3.chr3L 17219157 94 - 24543557 -GAAAGGCAGACAGAGAUAGCAUUAUAUGCAAAUGUA-AUGCAGAAGCGAGA-GGGCA--ACACUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU -.....(((.(..(((...(((((((((....)))))-))))....((....-..)).--...)))..).)))...((((((((....))))))))... ( -25.60, z-score = -3.74, R) >droSim1.chr3L 16554146 94 - 22553184 -GAAAGGCAGAUAGAGAUAGCAUUAUAUGCAAAUGUA-AUGCUGAAGCGAGA-GGCCA--ACACUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU -.....(((.((.(((.(((((((((((....)))))-))))))..((....-.))..--...))).)).)))...((((((((....))))))))... ( -27.20, z-score = -3.88, R) >droSec1.super_0 9271250 94 - 21120651 -GAAAGGCAAAUAGAGAUAGCAUUAUAUGCAAAUGUA-AUGCAGAAGCGAGA-GGGCA--ACACUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU -.....(((....(((...(((((((((....)))))-))))....((....-..)).--...)))....)))...((((((((....))))))))... ( -25.20, z-score = -3.79, R) >droYak2.chr3L 7342590 94 + 24197627 -GAAAGGCAGACAGAGAUGGUAUUAUAUGCAAAUGUA-AUGCAGAAGCGAGA-GGGCA--ACACUUGAUGUGCACACAAAGUCUGAACGGAUUUUGAAU -.....(((.(..(((.(((((((((((....)))))-))))....((....-..)).--.)))))..).)))...((((((((....))))))))... ( -24.10, z-score = -2.94, R) >droEre2.scaffold_4784 9131743 94 + 25762168 -GAAAGGCAGACAGAGAUAGUAUCAUAUGCAAAUCUA-AUGCAGAAGCGAGA-AGGCA--ACACUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU -((...(((....((.......))...)))...))..-(((((.(..((((.-.(...--.).)))).).))))).((((((((....))))))))... ( -19.40, z-score = -1.14, R) >droAna3.scaffold_13337 7341488 94 - 23293914 -GAAAGGCGGACAGAGAUAGUAUUAUAUGCAAAUGUG-CGGCAGAAGCGAGA-GGGCG--ACACUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU -....(.((.(((......((((...))))...))).-)).)....(((.((-((...--.).)))....)))...((((((((....))))))))... ( -19.60, z-score = -0.89, R) >droWil1.scaffold_181009 426930 95 + 3585778 GAAAGAGAUAGCAUAAAUGAUAU-GCAAAUGAAUGUG-G-GUGGAAGUGAGAUAGGCA-CACAUUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU ....................(((-(((..((((((((-.-((.............)).-))))))))...))))))((((((((....))))))))... ( -20.32, z-score = -2.27, R) >droMoj3.scaffold_6680 21500851 84 + 24764193 -------------UGGAU-GCAU-GUGUGUGAGUGCGUGUGUGCAAGCGAGAGAGCAACAAUUUUCUAUGCGCAUAGAAAGUCUGAACGGAUUUUGAAU -------------((.((-((((-(..(....)..))))))).)).(((..((((........))))...)))....(((((((....))))))).... ( -21.70, z-score = -0.48, R) >consensus _GAAAGGCAGACAGAGAUAGCAUUAUAUGCAAAUGUA_AUGCAGAAGCGAGA_GGGCA__ACACUUGAUGUGCAUACAAAGUCUGAACGGAUUUUGAAU ...................((((....(((....))).))))....(((....((........)).....)))...((((((((....))))))))... (-11.30 = -10.18 + -1.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:10 2011