| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,212,284 – 17,212,420 |
| Length | 136 |
| Max. P | 0.651090 |

| Location | 17,212,284 – 17,212,420 |
|---|---|
| Length | 136 |
| Sequences | 11 |
| Columns | 158 |
| Reading direction | reverse |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.56445 |
| G+C content | 0.50623 |
| Mean single sequence MFE | -39.51 |
| Consensus MFE | -19.35 |
| Energy contribution | -19.04 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
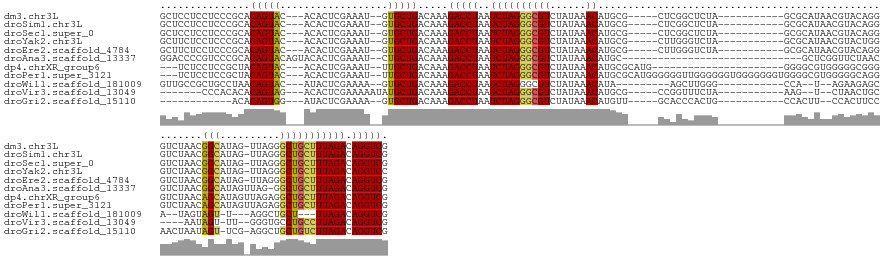
>dm3.chr3L 17212284 136 - 24543557 GCUCCUCCUCCCGCACAGUAC---ACACUCGAAAU--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG-----CUCGGCUCUA-----------GCGCAUAACGUACAGGGUCUAACGGCAUAG-UUAGGGCUGCUUUAGACAGGUCG ...............((((((---(.........)--)))))).....(((((..((((((((..........(((((-----((.......)-----------))))))........((.((((((......)-))))).)))))))))).))))). ( -42.40, z-score = -1.41, R) >droSim1.chr3L 16547354 136 - 22553184 GCUCCUCCUCCCGCACAGUAC---ACACUCGAAAU--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG-----CUCGGCUCUA-----------GCGCAUAACGUACAGGGUCUAACGGCAUAG-UUAGGGCUGCUUUAGACAGGUCG ...............((((((---(.........)--)))))).....(((((..((((((((..........(((((-----((.......)-----------))))))........((.((((((......)-))))).)))))))))).))))). ( -42.40, z-score = -1.41, R) >droSec1.super_0 9264492 136 - 21120651 GCUCCUCCUCCCGCACAGUAC---ACACUCGAAAU--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG-----CUCGGCUCUA-----------GCGCAUAACGUACAGGGUCUAACGGCAUAG-UUAGGGCUGCUUUAGACAGGUCG ...............((((((---(.........)--)))))).....(((((..((((((((..........(((((-----((.......)-----------))))))........((.((((((......)-))))).)))))))))).))))). ( -42.40, z-score = -1.41, R) >droYak2.chr3L 7335325 136 + 24197627 GCUUCUCCUCCCGCACAGUAC---ACACUCGAAAU--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG-----CUUGGGUCUA-----------GCGCAUAACGUACUGGGUCUAACGGCAUAG-UUAGGGCUGCUUUAGACAGGUCC ...............((((((---(.........)--)))))).....(((((..((((((((((((.(((..((((.-----.((((..(((-----------(..(.....)..))))..))))..))))..-))))))).)))))))).))))). ( -44.70, z-score = -2.00, R) >droEre2.scaffold_4784 9124758 136 + 25762168 GCUUCUCCUCCCGCACAGUAC---ACACUCGAAAU--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG-----CUUGGGUCUA-----------GCGCAUAACGUACAGGGUCUAACGGCAUAG-UUAGGGCUGCUUUAGACAGGUCG ...............((((((---(.........)--)))))).....(((((..((((((((..........(((((-----((.......)-----------))))))........((.((((((......)-))))).)))))))))).))))). ( -43.20, z-score = -1.69, R) >droAna3.scaffold_13337 7335176 125 - 23293914 GGACCCCGUCCCGCACAGUACAGUACACUCGAAAU--CUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGC------------------------------GCUCGGUUCUAACGUCUAACGGCAUAGUUAG-GGCUGCUUUAGACAGGUCG ((((...))))......((.(((((..........--.)))))))...(((((..((((((((((..........))------------------------------))))((((((((((((....)))...)))))-))))....)))).))))). ( -36.30, z-score = -1.09, R) >dp4.chrXR_group6 379312 128 + 13314419 ---UCUCCUCCGCUACAGUAC---ACACUCGAAAU--UUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCAUG----------------------GGGGCGUGGGGGCGGGGUCUAACAGCAUAGUUAGAGGCUGCUUUAGACAGGUCG ---............(((((.---...........--.))))).....(((((..((((((((((((.....(((((.(...----------------------.).))))).))))((..((((((......))))))..)))))))))).))))). ( -38.22, z-score = -0.33, R) >droPer1.super_3121 3344 150 + 4875 ---UCUCCUCCGCUACAGUAC---ACACUCGAAAU--UUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCGCAUGGGGGGUUGGGGGGUGGGGGGGUGGGGCGUGGGGGCAGGGUCUAACAGCAUAGUUAGAGGCUGCUUUAGACAGGUCG ---.((((((((((.(((((.---...........--.))))).......((((((((...((((..........)))).....)))))))).))))))))))...(((.((.((((((..((((((......))))))..))))))....)).))). ( -53.32, z-score = -1.64, R) >droWil1.scaffold_181009 3169157 120 - 3585778 GUUGCCGCUGCCUAACAGUAC---AUACUCGAAAA--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUA---------AGCUUGGG-----------CCA--U--AGAAGAGCA--UAGUAGU-U---AGGCUGCU---UUAGACAGGUCG ...............((((((---...........--)))))).....(((((..(((((((((((((...((.((---------.((((...-----------(..--.--.)..)))).--))))...-)---)))).)))---))))).))))). ( -32.30, z-score = -0.30, R) >droVir3.scaffold_13049 20347932 121 + 25233164 -------CCCACACACAGUAG---ACACUCGAAAAAUAUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG-----CCGGUUUCUA-----------AAG--U--CUAACUGC----AAUAGU-UU--GGGUGCCUGCCUUAGACAGGUCG -------........(((((.---..............))))).....(((((..((((((((((......))..(((-----((.(...(((-----------..(--(--......))----..))).-.)--.)))))..)))))))).))))). ( -32.16, z-score = -1.46, R) >droGri2.scaffold_15110 10051682 121 - 24565398 ------------ACACAGUGG---AUACUCGAAAA--GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGUU-----GCACCCACUG-----------CCACUU--CCACUUCCAACUAAUAGU-UCG-AGGCUGCUGUCUUAGACAGGUCG ------------.....((.(---.((((.....)--))).).))...(((((..((((((((............((.-----(((.....))-----------).))..--.((((((.(((.....))-).)-))).))..)))))))).))))). ( -27.20, z-score = 0.12, R) >consensus GCUCCUCCUCCCGCACAGUAC___ACACUCGAAAU__GUGCUGACAAAGACCUAAUCUAGGGCGUCUAUAAACAUGCG_____CUCGGCUCUA___________GCGCAUAACGUACAGGGUCUAACGGCAUAG_UUGGGGCUGCUUUAGACAGGUCG ...............(((((..................))))).....(((((..((((((((((......))......................................................(((..........))))))))))).))))). (-19.35 = -19.04 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:09 2011