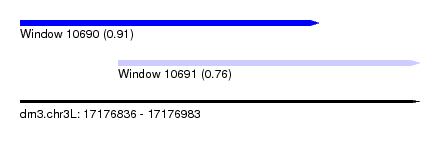
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,176,836 – 17,176,983 |
| Length | 147 |
| Max. P | 0.911458 |

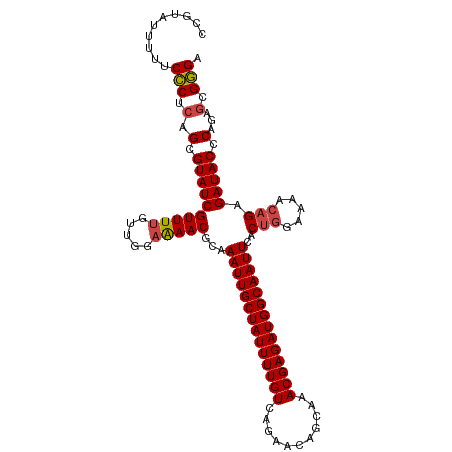
| Location | 17,176,836 – 17,176,946 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Shannon entropy | 0.21272 |
| G+C content | 0.43288 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -23.73 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17176836 110 + 24543557 CCGUAUUUUUCCCUCAGCGUAUCGUUUUGUUGGAAAACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGA ........(((((((.(.(((((.....(((....)))...((((((((((((((..(.....)..))))))))))))))..(((.....))).))))).).))).)))) ( -30.20, z-score = -1.87, R) >droEre2.scaffold_4784 9087949 110 - 25762168 CUUUAUUUUUCCCUCAGCGUAUCGUUUUGUUGGAAAACGCAAAUUGCUAUUUUGUCUGAACAGCAAACGAGAUGGCAAUUCACUGGAAAGCAGAGAUACCCAGAGCGGGG ..........(((((((((........))))))....(((.((((((((((((((.((.....)).))))))))))))))..((((.............)))).)))))) ( -31.42, z-score = -1.93, R) >droYak2.chr3L 7298849 109 - 24197627 CUUUAUUUU-CCCUCAGCGUAUCGUUUUGUUGGAAAACGCAAAUUGCUAUUUUGUCUGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGC ........(-(((((.(.(((((.....(((....)))...((((((((((((((.((.....)).))))))))))))))..(((.....))).))))).).))).))). ( -31.40, z-score = -2.36, R) >droSec1.super_0 9228784 109 + 21120651 CCGUAUUUU-CCCACAGCGUAUCGUUUUGUUGGAAAACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGA ((...((((-((.((((((...)))..))).))))))(((.((((((((((((((..(.....)..))))))))))))))..((((.............)))).))))). ( -30.92, z-score = -2.42, R) >droSim1.chr3L 16511179 110 + 22553184 CAGUAUUUUUCCCUCAGCGUAUCGUUUUGUUGGAAAACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGA ........(((((((.(.(((((.....(((....)))...((((((((((((((..(.....)..))))))))))))))..(((.....))).))))).).))).)))) ( -30.20, z-score = -2.07, R) >droAna3.scaffold_13337 7300244 101 + 23293914 --------UUCUCCCAGCGUAUCGUUUC-UUGUAGAACGCAAAUUGCUAUUUUGUCUGAACAGAAAACGAGAUGGCAAUUCACAAAACGAAAGAGAUACCCGGCCAGAGA --------.((((((.(.(((((((((.-.....))))(..((((((((((((((((....)))...)))))))))))))..)....(....).)))))).))...)))) ( -26.30, z-score = -2.36, R) >consensus CCGUAUUUUUCCCUCAGCGUAUCGUUUUGUUGGAAAACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGA ..........(((.(.(.((((((((((.....)))))...((((((((((((((...........))))))))))))))..(((.....))).))))).)...).))). (-23.73 = -23.98 + 0.25)

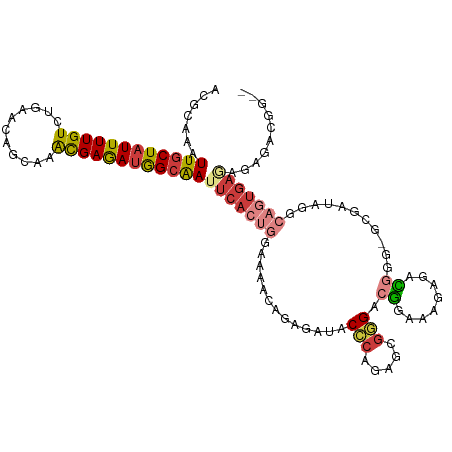
| Location | 17,176,872 – 17,176,983 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Shannon entropy | 0.42617 |
| G+C content | 0.48107 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -15.06 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17176872 111 + 24543557 ACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGACGGAAAGAGACGGG-GCGAUAGGCAGUGAUAGAGACGG-- .(((..(((((((((((((..(.....)..)))))))))))))((((((.............(((.....))).((........))..-........))))))....).)).-- ( -26.80, z-score = -2.28, R) >droEre2.scaffold_4784 9087985 111 - 25762168 ACGCAAAUUGCUAUUUUGUCUGAACAGCAAACGAGAUGGCAAUUCACUGGAAAGCAGAGAUACCCAGAGCGGGGCGGAAAGAGAUGGG-GCGACAGGCAGUGAGAGAGACGG-- .(((...((((((((((((.((.....)).))))))))))))(((((((.............(((.....)))..............(-....)...)))))))...).)).-- ( -28.30, z-score = -1.28, R) >droYak2.chr3L 7298884 111 - 24197627 ACGCAAAUUGCUAUUUUGUCUGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGCCGGAAAGAGAUGGG-GCGACAGGCAGUGAGAGAGACGG-- .(((...((((((((((((.((.....)).))))))))))))(((((((.............(((.....)))(((........))).-........)))))))...).)).-- ( -28.90, z-score = -1.56, R) >droSec1.super_0 9228819 111 + 21120651 ACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGACGGAAAGGGACGGG-GCGAUAGGCAGUGAGAGAGACGG-- .(((...((((((((((((..(.....)..))))))))))))(((((((.............(((.....))).((........))..-........)))))))...).)).-- ( -27.10, z-score = -2.06, R) >droSim1.chr3L 16511215 111 + 22553184 ACGCAAAUUGCUAUUUUGUCAGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGACGGAAAGGGACGGG-GCGAUAGGCAGUGAGAGAGACGG-- .(((...((((((((((((..(.....)..))))))))))))(((((((.............(((.....))).((........))..-........)))))))...).)).-- ( -27.10, z-score = -2.06, R) >droWil1.scaffold_181009 372597 107 - 3585778 ACGCAAAUUGCUAUUUUGUUUGAGUCACAAGUGAAAAAGAGA-----AAGAACAACCAAACACAAAGAAUAUGAGCGAAAGAGAGAGACUCUAUGGAGGACAGCAGAGAGGG-- .......(((((.((((.(..(((((....(((.........-----.............)))............(....).....)))))...).)))).)))))......-- ( -15.55, z-score = -0.08, R) >droAna3.scaffold_13337 7300271 113 + 23293914 ACGCAAAUUGCUAUUUUGUCUGAACAGAAAACGAGAUGGCAAUUCACAAAACGAAAGAGAUACCCGGCCAGAGAGGAAGUGAGACAGA-UUAACAUAGAAUGAAAGAGAUGGGU ..(..((((((((((((((((....)))...)))))))))))))..)....(....)....(((((.((.....))..((........-...))...............))))) ( -20.70, z-score = -1.46, R) >consensus ACGCAAAUUGCUAUUUUGUCUGAACAGCAAACGAGAUGGCAAUUCACUGGAAAACAGAGAUACCCAGAGCGGGACGGAAAGAGACGGG_GCGAUAGGCAGUGAGAGAGACGG__ ......(((((((((((((...........)))))))))))))((((((.............(((.....))).((........))...........))))))........... (-15.06 = -16.13 + 1.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:07 2011