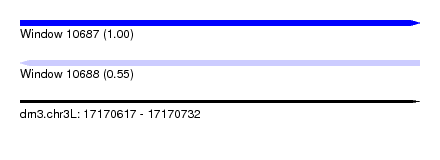
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,170,617 – 17,170,732 |
| Length | 115 |
| Max. P | 0.997459 |

| Location | 17,170,617 – 17,170,732 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.37 |
| Shannon entropy | 0.05520 |
| G+C content | 0.27790 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -25.02 |
| Energy contribution | -25.11 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.997459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17170617 115 + 24543557 -----UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA -----....(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.64, R) >droAna3.scaffold_13337 7293961 113 + 23293914 ------UUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUU-AUUA ------...(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........)))))))))))))).-.... ( -26.00, z-score = -3.64, R) >droEre2.scaffold_4784 9081545 115 - 25762168 -----UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA -----....(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.64, R) >droYak2.chr3L 7292383 115 - 24197627 -----UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA -----....(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.64, R) >droSec1.super_0 9222596 115 + 21120651 -----UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGAGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA -----....(((((((((.(((((.........((((.....(((.((((((..((((.....)))).)))))))))...)))).((((......)))).))))))))))))))...... ( -22.00, z-score = -2.11, R) >droSim1.chr2h_random 1115134 115 - 3178526 -----UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA -----....(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.64, R) >droWil1.scaffold_181009 365928 115 - 3585778 -----UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA -----....(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.64, R) >droMoj3.scaffold_6680 21441438 114 - 24764193 ------UUUGAAUUUUGUUGUCGCACCUUUUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA ------...(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.70, R) >droGri2.scaffold_15110 10008201 114 + 24565398 ------UUUGAAUUUUGUUGUCGCACCUUUUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA ------...(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.70, R) >droVir3.scaffold_13049 20298507 114 - 25233164 ------UUUGAAUUUUGUUGUCGCACCUUUUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA ------...(((((((((.(((((.(((....))).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.70, R) >droPer1.super_1156 3961 120 + 8443 GUGUGUUUUGAAUUUUGUUGUCGCACCUUCUCUGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA .........(((((((((.(((((.((......)).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -2.81, R) >dp4.chrXR_group6 13311140 120 + 13314419 GUGUAUUUUGAAUUUUGUUGUCGCACCUUCUCUGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA .........(((((((((.(((((.((......)).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... ( -26.00, z-score = -3.12, R) >consensus _____UUUUGAAUUUUGUUGUCGCACCUUCUCAGGAAUUCUGUUUACAUUUAUUACUUUGCGCAAGUUUAAGUGAAAAAUUUCCAUAAACAUUCUUUUAAGCGACAUAAAGUUUUUAUUA .........(((((((((.(((((.((......)).....((((((.((((.((((((.((....))..)))))).)))).....)))))).........))))))))))))))...... (-25.02 = -25.11 + 0.08)

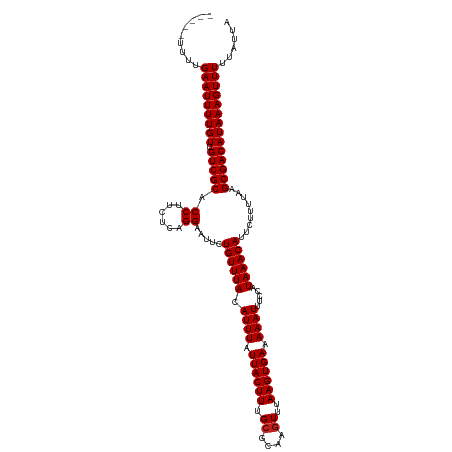
| Location | 17,170,617 – 17,170,732 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.37 |
| Shannon entropy | 0.05520 |
| G+C content | 0.27790 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -17.62 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.549389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17170617 115 - 24543557 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA----- ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).............----- ( -18.00, z-score = -0.80, R) >droAna3.scaffold_13337 7293961 113 - 23293914 UAAU-AAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAA------ ....-.........((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).))))))............------ ( -18.00, z-score = -0.74, R) >droEre2.scaffold_4784 9081545 115 + 25762168 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA----- ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).............----- ( -18.00, z-score = -0.80, R) >droYak2.chr3L 7292383 115 + 24197627 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA----- ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).............----- ( -18.00, z-score = -0.80, R) >droSec1.super_0 9222596 115 - 21120651 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCUCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA----- ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).............----- ( -18.00, z-score = -1.05, R) >droSim1.chr2h_random 1115134 115 + 3178526 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA----- ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).............----- ( -18.00, z-score = -0.80, R) >droWil1.scaffold_181009 365928 115 + 3585778 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA----- ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).............----- ( -18.00, z-score = -0.80, R) >droMoj3.scaffold_6680 21441438 114 + 24764193 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAAAAGGUGCGACAACAAAAUUCAAA------ ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).))))))............------ ( -19.50, z-score = -1.61, R) >droGri2.scaffold_15110 10008201 114 - 24565398 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAAAAGGUGCGACAACAAAAUUCAAA------ ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).))))))............------ ( -19.50, z-score = -1.61, R) >droVir3.scaffold_13049 20298507 114 + 25233164 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAAAAGGUGCGACAACAAAAUUCAAA------ ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).))))))............------ ( -19.50, z-score = -1.61, R) >droPer1.super_1156 3961 120 - 8443 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCAGAGAAGGUGCGACAACAAAAUUCAAAACACAC ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....((......)).)))))).................. ( -16.20, z-score = -0.19, R) >dp4.chrXR_group6 13311140 120 - 13314419 UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCAGAGAAGGUGCGACAACAAAAUUCAAAAUACAC ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....((......)).)))))).................. ( -16.20, z-score = -0.23, R) >consensus UAAUAAAAACUUUAUGUCGCUUAAAAGAAUGUUUAUGGAAAUUUUUCACUUAAACUUGCGCAAAGUAAUAAAUGUAAACAGAAUUCCUGAGAAGGUGCGACAACAAAAUUCAAAA_____ ..............((((((.........(((((((.(((....)))......((((.....)))).......))))))).....(((....))).)))))).................. (-17.62 = -17.79 + 0.17)
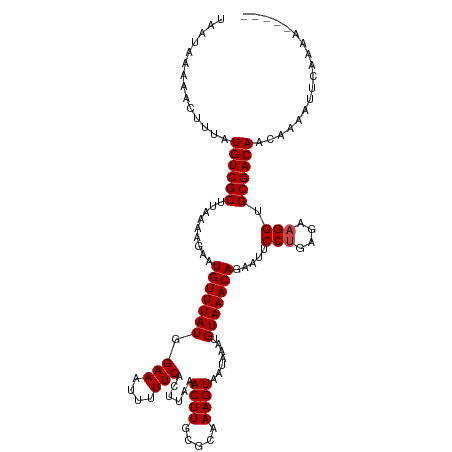
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:04 2011