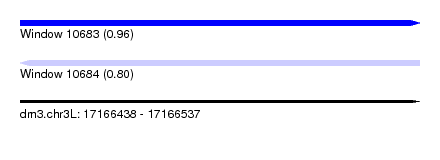
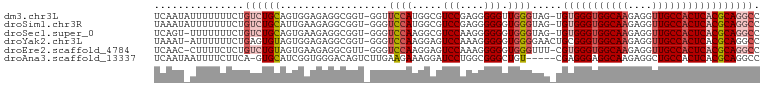
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,166,438 – 17,166,537 |
| Length | 99 |
| Max. P | 0.958265 |

| Location | 17,166,438 – 17,166,537 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Shannon entropy | 0.38936 |
| G+C content | 0.57137 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -17.39 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
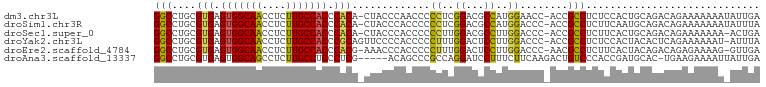
>dm3.chr3L 17166438 99 + 24543557 GGCCUGCGUGAGUGGCAACCUCUUGCCACCCACA-CUACCCAACCCCCUCGGACGCCAUGGAACC-ACCGCCUCUCCACUGCAGACAGAAAAAAAUAUUGA ...(((((((.(((((((....))))))).))).-...............(((.((..((....)-)..))...)))...))))................. ( -25.80, z-score = -2.55, R) >droSim1.chr3R 9413661 99 + 27517382 GGCCUGCGUGAGUGGCAACCUCUUGCCACCCACA-CUACCCACCCCCCUCGGACGCCAUGGACCC-ACCGCCUCUUCAAUGCAGACAGAAAAAAAUAUUUA ...(((((((.(((((((....))))))).))).-...............(((.((..((....)-)..)).))).....))))................. ( -24.80, z-score = -2.46, R) >droSec1.super_0 9218451 98 + 21120651 GGCCUGCGUGAGUGGCAACCUCUUGCCACCCACA-CUACCCACCCCCCUUGGACGCCUUGGACCC-ACCGCCUCUUCACUGCAGACAGAAAAAAA-ACUGA ...(((((((.(((((((....))))))).))).-...............(((.((..((....)-)..)).))).....)))).(((.......-.))). ( -26.10, z-score = -2.48, R) >droYak2.chr3L 7288239 99 - 24197627 GGCCUGCGUGAGUGGCAACCUCUUGCCACCCGCAGUUCCCCACCCCCUUUGGACUCCUUGGACCC-ACCGCCUCUCCACUACACUCAGAAAAAAU-AUUUA ((((((((.(.(((((((....)))))))))))))..............(((..((....)).))-)..))).......................-..... ( -25.20, z-score = -2.69, R) >droEre2.scaffold_4784 9077849 98 - 25762168 GGCCUGCGUGAGUGGCAACCUCUUGCCACCCACG-AAACCCACCCCCUUUGGACUCCUUGGACCC-AACGCCUCUUCACUACAGACAGAGAAAAG-GUUGA ((....((((.(((((((....))))))).))))-....))....(((((((..((....)).))-.....((((((......)).)))).))))-).... ( -28.10, z-score = -1.68, R) >droAna3.scaffold_13337 7289691 95 + 23293914 GGCCUGCGUGAGUGGCAGCCUCUUGCCUCCCUCG-----ACAGCCCGCCAGGAUCCUUUCUUCAAGACUGUCCCACCGAUGCAC-UGAAGAAAAUUAUUGA (((..((.((((.(((((....)))))...))))-----...))..))).......((((((((.....(((.....)))....-))))))))........ ( -23.90, z-score = -0.30, R) >consensus GGCCUGCGUGAGUGGCAACCUCUUGCCACCCACA_CUACCCACCCCCCUUGGACGCCUUGGACCC_ACCGCCUCUCCACUGCAGACAGAAAAAAA_AUUGA (((....(((.(((((((....))))))).))).............((..((...))..))........)))............................. (-17.39 = -16.92 + -0.47)
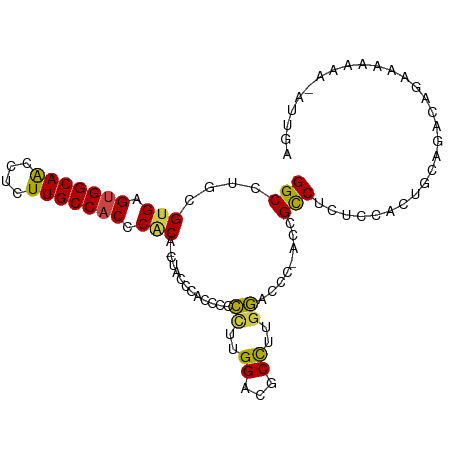
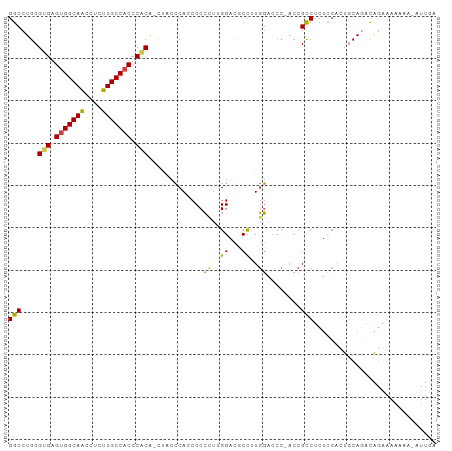
| Location | 17,166,438 – 17,166,537 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.42 |
| Shannon entropy | 0.38936 |
| G+C content | 0.57137 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
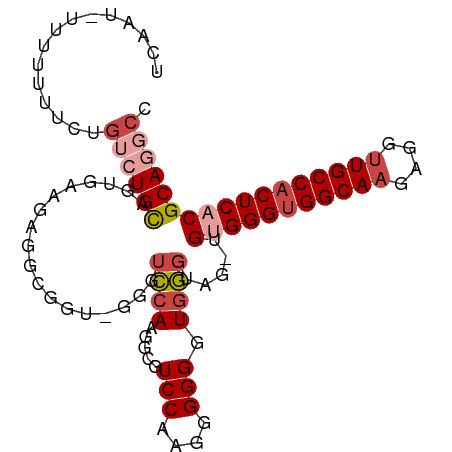
>dm3.chr3L 17166438 99 - 24543557 UCAAUAUUUUUUUCUGUCUGCAGUGGAGAGGCGGU-GGUUCCAUGGCGUCCGAGGGGGUUGGGUAG-UGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCC .((((.(((((..(((....)))..)))))((.((-(....))).))..((....))))))(((.(-((((((((((((....)))))))))))))..))) ( -37.90, z-score = -2.05, R) >droSim1.chr3R 9413661 99 - 27517382 UAAAUAUUUUUUUCUGUCUGCAUUGAAGAGGCGGU-GGGUCCAUGGCGUCCGAGGGGGGUGGGUAG-UGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCC ............((..(((.(.(((..((.((.((-(....))).)).))))).).)))..))..(-((((((((((((....)))))))))))))..... ( -39.20, z-score = -2.83, R) >droSec1.super_0 9218451 98 - 21120651 UCAGU-UUUUUUUCUGUCUGCAGUGAAGAGGCGGU-GGGUCCAAGGCGUCCAAGGGGGGUGGGUAG-UGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCC ...((-((((((.(((....))).))))))))...-.((((....((.(((...))).)).....(-((((((((((((....))))))))))))).)))) ( -38.50, z-score = -2.33, R) >droYak2.chr3L 7288239 99 + 24197627 UAAAU-AUUUUUUCUGAGUGUAGUGGAGAGGCGGU-GGGUCCAAGGAGUCCAAAGGGGGUGGGGAACUGCGGGUGGCAAGAGGUUGCCACUCACGCAGGCC ...((-((((.....))))))...((....((((.-.(.(((....(.(((....))).)..))).)..)(((((((((....))))))))).)))...)) ( -30.50, z-score = -0.62, R) >droEre2.scaffold_4784 9077849 98 + 25762168 UCAAC-CUUUUCUCUGUCUGUAGUGAAGAGGCGUU-GGGUCCAAGGAGUCCAAAGGGGGUGGGUUU-CGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCC ....(-((((((.(((....))).)))))))..((-(((((....)).)))))........(((((-((((((((((((....)))))))))))).))))) ( -39.70, z-score = -2.26, R) >droAna3.scaffold_13337 7289691 95 - 23293914 UCAAUAAUUUUCUUCA-GUGCAUCGGUGGGACAGUCUUGAAGAAAGGAUCCUGGCGGGCUGU-----CGAGGGAGGCAAGAGGCUGCCACUCACGCAGGCC .......(((((((((-(.((.((.....))..)).)))))))))).........((.(((.-----((.(((.((((......)))).))).))))).)) ( -28.40, z-score = 0.41, R) >consensus UCAAU_UUUUUUUCUGUCUGCAGUGAAGAGGCGGU_GGGUCCAAGGCGUCCAAGGGGGGUGGGUAG_UGUGGGUGGCAAGAGGUUGCCACUCACGCAGGCC ...............((((((..................((((.....(((....))).)))).....(((((((((((....))))))))))))))))). (-23.26 = -24.60 + 1.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:33:01 2011