
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,120,001 – 17,120,094 |
| Length | 93 |
| Max. P | 0.720828 |

| Location | 17,120,001 – 17,120,094 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.43 |
| Shannon entropy | 0.47210 |
| G+C content | 0.53089 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.87 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
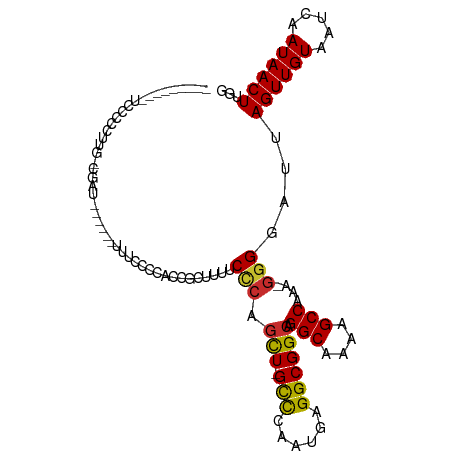
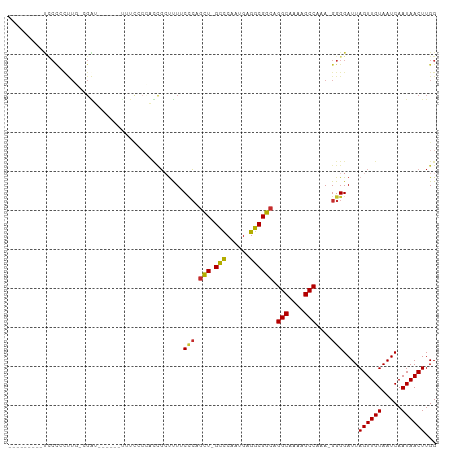
>dm3.chr3L 17120001 93 + 24543557 ---------UCCCCCUUGGCGAU------UUUCACCAUCGCUUUUCCCAGCU-GCCCAAUGAGGCGGCAGGCAAAAGCCAAA-GGGGAUUAGUUGUAAUCAAUAACUUGG ---------..((((((((.(..------...).))...((((((.((.(((-(((......)))))).)).))))))..))-))))...((((((.....))))))... ( -34.00, z-score = -2.70, R) >droSim1.chr3L 16455428 94 + 22553184 --------CUCCCGCUUGGCGAU------UUUCCCCACCGCUUUUCCCAGCU-GCCCAAUGAGGCGGCAGGCAAAAGCCAAA-GGGGAUUAGUUGUAAUCAAUAACUUGG --------..........(((((------(.(((((...((((((.((.(((-(((......)))))).)).))))))....-)))))..)))))).............. ( -33.20, z-score = -1.86, R) >droSec1.super_0 9170791 94 + 21120651 --------CUCCCGCUUGGCGAU------UUUCCCCACCGCUUUUCCCAGCU-GCCCAAUGAGGCGGCAGGCAAAAGCCAAA-GGGGAUUAGUUGUAAUCAAUAACUUGG --------..........(((((------(.(((((...((((((.((.(((-(((......)))))).)).))))))....-)))))..)))))).............. ( -33.20, z-score = -1.86, R) >droYak2.chr3L 7239126 89 - 24197627 ---------UUCCCUUU--CGAU------UUUCCC--UCGGUUUUCCCAGCU-GCCCAAUGAGGCGGCAGGCAAGAGCCAAA-GGGGAUUAGUUGUAAUCAAUAACUUGG ---------((((((((--.((.------......--))((((((.((.(((-(((......)))))).))..)))))))))-)))))..((((((.....))))))... ( -28.50, z-score = -1.45, R) >droEre2.scaffold_4784 9030359 89 - 25762168 ---------UGCUCUUU--CGAU------UUUCCC--UCGGCUUUCCCAGCU-GCCCAAUGAGGCGGCAGGCAAAAGCCAAA-GGGGAUUAGUUGUAAUCAAUAACUUGG ---------........--....------.(((((--(.((((((.((.(((-(((......)))))).))..))))))..)-)))))..((((((.....))))))... ( -30.00, z-score = -1.64, R) >droAna3.scaffold_13337 7243627 93 + 23293914 --------UUCCCCCCU--CGGU------CCACCCCGUCCCUUUUCAGAUUUUGCCCAAUGAGGCGGCAGGCGAAAGCCAAA-GGGGAUUAGUUGUAAUCAAUAACUUGG --------.........--.((.------...))(((((((((((.......(((((......).))))(((....))))))-)))))).((((((.....)))))).)) ( -26.80, z-score = -0.70, R) >dp4.chrXR_group6 13248782 97 + 13314419 CCCAGGGCCCUCCCCCUGCGGGU------------UGCUGCUCUGCUCUGCU-GCCCAAUGAGGCGGCAGGCGAAAGCCAAAAGGGGAUUAGUUGUAAUCAAUAACUUGG .(((((......(((((((((((------------....)).))))..((((-(((......)))))))(((....)))...)))))....((((....))))..))))) ( -40.50, z-score = -1.42, R) >droPer1.super_250 11901 109 + 39459 CCCAGGGCCCUCCCCCUGCGGGUGGGUGCUGCUGCUGCUGCUCUGCUCUGCU-GCCCAAUGAGGCGGCAGGCGAAAGCCAAAAGGGGAUUAGUUGUAAUCAAUAACUUGG .(((((......(((((..(((..((.((.((....)).))))..)))((((-(((......)))))))(((....)))...)))))....((((....))))..))))) ( -47.30, z-score = -1.15, R) >droGri2.scaffold_15110 9955373 72 + 24565398 ------------------------------------CAAGCCCUGCUC-GCU-GUUCAAUGAGGCGGCAGGCAGAAGCCAAAAGGGGAUUAGUUGUAAUCAAUAACUUGG ------------------------------------((((((((((.(-(((-.........)))))).(((....)))...))))(((((....))))).....)))). ( -22.20, z-score = -1.28, R) >consensus _________UCCCCCUUG_CGAU______UUUCCCCACCGCUUUUCCCAGCU_GCCCAAUGAGGCGGCAGGCAAAAGCCAAA_GGGGAUUAGUUGUAAUCAAUAACUUGG .............................................(((.(((.(((......)))))).(((....)))....)))....((((((.....))))))... (-17.01 = -16.87 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:55 2011