| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,070,044 – 17,070,134 |
| Length | 90 |
| Max. P | 0.976292 |

| Location | 17,070,044 – 17,070,134 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.44 |
| Shannon entropy | 0.55128 |
| G+C content | 0.70774 |
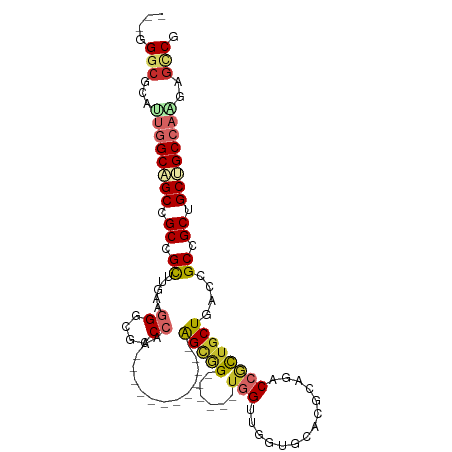
| Mean single sequence MFE | -50.34 |
| Consensus MFE | -25.47 |
| Energy contribution | -26.52 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976292 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
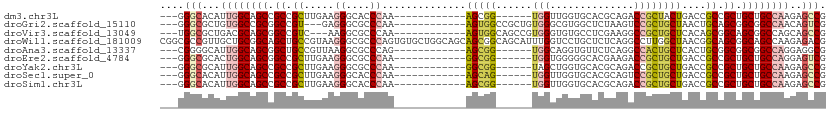
>dm3.chr3L 17070044 90 + 24543557 ---GGGCACAUUGGCAGCCGCCGCUUGAAGGGCACCCAA------------AGCGG------UGGUUGGUGCACGCAGACCGCUACUGACCGCCGCUGCUGCCAAGAGCCG ---.(((...((((((((.(((........)))......------------(((((------((((..((((.........)).))..)))))))))))))))))..))). ( -48.30, z-score = -3.23, R) >droGri2.scaffold_15110 9913227 93 + 24565398 ---GGGCGCUGUGGCCGCGGCCGU---GAGGGCGCCCAA------------AGUGGCCGCUGUGGGCGUGGCUCUAAGUCCGCUGCUAACUGCAGCGGCGGCCAACAGUCG ---....((((((((((((((...---(((.(((((((.------------(((....))).)))))))..)))...)))((((((.....)))))))))))).))))).. ( -52.20, z-score = -1.73, R) >droVir3.scaffold_13049 20194572 93 - 25233164 ---UGGCGCUGACGCAGCGGCCGUC---AAGGCGCCCAA------------AGUGGCAGCCGUGGGUGUGCCUCGAAGGCCGCUGCUCACAGCGGCAGCGGCCAGCAGCCG ---.(((((((..(((((((((.((---.(((((((((.------------.((....))..)))))..)))).)).)))))))))...))))(((....)))....))). ( -54.80, z-score = -2.45, R) >droWil1.scaffold_181009 255918 111 - 3585778 CGGCGCCGUUGCUGCGGCAGCUGCCGUAAGGGCGCCCAGUGUGCUGGCAGCAGCGGCAGCAUUUGGUCCUGCUCUCAGGCCUUGGCUAACGGCAGCGGCAGCCAAGAGACG .(((((((((((((.(((.((((((((....((..((((....))))..)).))))))))....((.((((....))))))...)))..)))))))))).)))........ ( -59.10, z-score = -1.29, R) >droAna3.scaffold_13337 7202105 90 + 23293914 ---CGGGGCAUUGGCAGCGGCUGCCGUUAAGGCGCCCAG------------AGCGG------UGGCAGGUGUUCUCAGGCCACUGCUCACUGCGGCGGCGGCCAGGAGGCG ---....((.(((((.((.((((((.....)))((...(------------(((((------((((..(......)..))))))))))...))))).)).)))))...)). ( -46.70, z-score = -1.32, R) >droEre2.scaffold_4784 8989568 90 - 25762168 ---GGGCGCACUGGCAGCGGCCGCUUGAAGGGCGCCCAA------------GGCGG------UGGUGGGGGCACGAAGACCGCUGCUGACCGCCGCUGCUGCCAGGAGUCG ---.(((...(((((((((((((((.....)))).....------------(((((------(((..(.((........)).)..)).)))))))))))))))))..))). ( -51.80, z-score = -2.09, R) >droYak2.chr3L 7196728 90 - 24197627 ---GGGCGCAUUGGCAGCCGCCGCUUGAAGGGCGCCCAA------------GGCGG------UAGCUGGUGCACGCAGACCGCUGCUGACCGCCGCUGCUGCCAAGAGCCG ---.(((...((((((((.((((((.....)))).....------------(((((------(.((....))..((((....))))..)))))))).))))))))..))). ( -47.00, z-score = -1.14, R) >droSec1.super_0 9130317 90 + 21120651 ---GGGCACAUUGGCAGCCGCCGCUUGAAGGGCACCCAA------------AGCAG------UGGUUGGUGCACGCAGUCCGCUGCUGACCGCCGCUGCUGCCAAGAGCCG ---.(((...((((((((....((((...((....)).)------------)))((------((((.(((.((.((((....)))))))))))))))))))))))..))). ( -45.30, z-score = -1.81, R) >droSim1.chr3L 16413053 90 + 22553184 ---GGGCACAUUGGCAGCCGCCGCUUGAAGGGCACCCAA------------AGCGG------UGGUUGGUGCACGCAGACCGCUGCUGACCGCCGCUGCUGCCAAGAGCCG ---.(((...((((((((.(((........)))......------------(((((------((((..((((.........)).))..)))))))))))))))))..))). ( -47.90, z-score = -2.51, R) >consensus ___GGGCGCAUUGGCAGCCGCCGCUUGAAGGGCGCCCAA____________AGCGG______UGGUUGGUGCACGCAGACCGCUGCUGACCGCCGCUGCUGCCAAGAGCCG ....(((...((((((((.((.((.....((....))..............(((((........((....)).......))))).......)).)).))))))))..))). (-25.47 = -26.52 + 1.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:48 2011