
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,067,737 – 17,067,837 |
| Length | 100 |
| Max. P | 0.793060 |

| Location | 17,067,737 – 17,067,837 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Shannon entropy | 0.28085 |
| G+C content | 0.37116 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.85 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
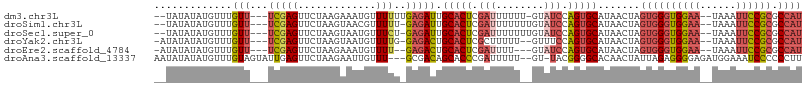
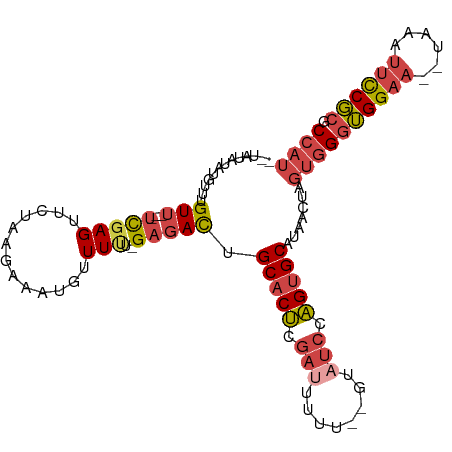
>dm3.chr3L 17067737 100 + 24543557 --UAUAUAUGUUUGUU---UCGAGUUCUAAGAAAUGUUUUUUGAGAUUGCACUCGAUUUUUU-GUAUCCAGUGCAUAACUAGUGGGUGGAA--UAAAUUCCGCGCCAU --...........(((---(((((.....(....)....))))))))((((((.(((.....-..))).))))))......((((((((((--....)))))).)))) ( -24.60, z-score = -2.11, R) >droSim1.chr3L 16410755 100 + 22553184 --UAUAUAUGUUUGUU---UCGAGUUCUAAGUAACGUUUUU-GAGAUUGCACUCGAUUUUUUUGUAUCCAGUGCAUAACUAGUGGGUGGAA--UAAAUUCCGCGCCAU --...........(((---(((((..(........)..)))-)))))((((((.(((........))).))))))......((((((((((--....)))))).)))) ( -25.70, z-score = -2.38, R) >droSec1.super_0 9128022 100 + 21120651 --UAUAUAUGUUUGUU---UCGAGUUCUAAGUAAUGUUUCU-GAGAUUGCACUCGAUUUUUUUGUAUCCAGUGCAUAACUAGUGGGUGGAA--UAAAUUCCGCGCCAU --...........(((---(((.(..(........)..).)-)))))((((((.(((........))).))))))......((((((((((--....)))))).)))) ( -22.70, z-score = -1.36, R) >droYak2.chr3L 7194399 99 - 24197627 -AUAUAUAUGUUUGUU---UCGAGUUCUAAGUAAUGUUUUG-GAGACUGCACUCGCUUUUU--GUUUCCAGUGCAUAACUAGUGGGUGGAA--UAAAUUCCGCGCCAU -...............---..........(((.((((.(((-(((((.((....)).....--)))))))).)))).))).((((((((((--....)))))).)))) ( -27.60, z-score = -2.49, R) >droEre2.scaffold_4784 8987379 97 - 25762168 -AUAUAUAUGUUUGUU---UCGAGUUCUAAGAAAUGUUUU--GAGACUGCACUCGAUUUU---GUAUCCAGUGCAUAACUAGUGGGUGGAA--UAAAUUCCGCGCCAU -............(((---(((((..(........).)))--)))))((((((.(((...---..))).))))))......((((((((((--....)))))).)))) ( -26.40, z-score = -2.52, R) >droAna3.scaffold_13337 7199665 102 + 23293914 AAUAUAUAUGUUUGUAGUAUUGAGUUCUAAGAAUUGUUU---GCGACAGCACCCGAUUUUU--GU-UACGGGGCACAACUAUUAGAGGGGAGAUGGAAAUCCCCCCUU ..............((((((((.(((((((((((((..(---((....)))..))))))))--..-...))))).))).)))))((((((.(((....))).)))))) ( -29.20, z-score = -2.27, R) >consensus __UAUAUAUGUUUGUU___UCGAGUUCUAAGAAAUGUUUUU_GAGACUGCACUCGAUUUUU__GUAUCCAGUGCAUAACUAGUGGGUGGAA__UAAAUUCCGCGCCAU ......................((((.........((((....)))).(((((.(((........))).)))))..)))).((((((((((......)))))).)))) (-14.79 = -15.85 + 1.06)

| Location | 17,067,737 – 17,067,837 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.52 |
| Shannon entropy | 0.28085 |
| G+C content | 0.37116 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -12.52 |
| Energy contribution | -13.63 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
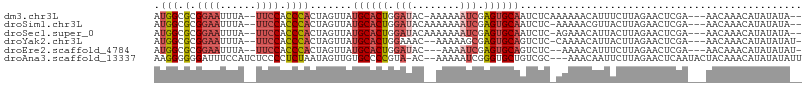
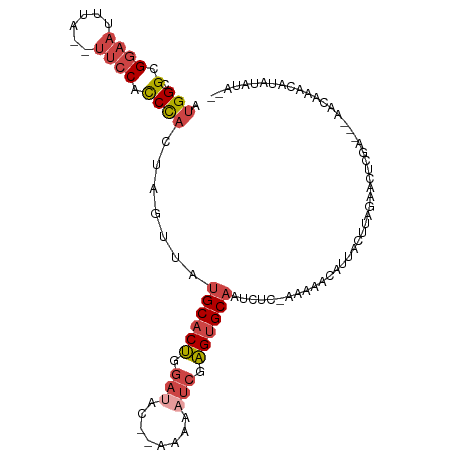
>dm3.chr3L 17067737 100 - 24543557 AUGGCGCGGAAUUUA--UUCCACCCACUAGUUAUGCACUGGAUAC-AAAAAAUCGAGUGCAAUCUCAAAAAACAUUUCUUAGAACUCGA---AACAAACAUAUAUA-- .(((.(.((((....--)))).))))...(((.((((((.(((..-.....))).)))))).............((((.........))---))..))).......-- ( -18.90, z-score = -2.48, R) >droSim1.chr3L 16410755 100 - 22553184 AUGGCGCGGAAUUUA--UUCCACCCACUAGUUAUGCACUGGAUACAAAAAAAUCGAGUGCAAUCUC-AAAAACGUUACUUAGAACUCGA---AACAAACAUAUAUA-- .(((.(.((((....--)))).))))...(((.((((((.(((........))).)))))).....-......(((......)))....---)))...........-- ( -19.50, z-score = -2.33, R) >droSec1.super_0 9128022 100 - 21120651 AUGGCGCGGAAUUUA--UUCCACCCACUAGUUAUGCACUGGAUACAAAAAAAUCGAGUGCAAUCUC-AGAAACAUUACUUAGAACUCGA---AACAAACAUAUAUA-- .(((.(.((((....--)))).))))...(((.((((((.(((........))).)))))).(((.-((........)).)))......---)))...........-- ( -20.70, z-score = -2.93, R) >droYak2.chr3L 7194399 99 + 24197627 AUGGCGCGGAAUUUA--UUCCACCCACUAGUUAUGCACUGGAAAC--AAAAAGCGAGUGCAGUCUC-CAAAACAUUACUUAGAACUCGA---AACAAACAUAUAUAU- .(((.(.((((....--)))).))))..((...((((((.(....--......).))))))..)).-......................---...............- ( -15.30, z-score = -0.36, R) >droEre2.scaffold_4784 8987379 97 + 25762168 AUGGCGCGGAAUUUA--UUCCACCCACUAGUUAUGCACUGGAUAC---AAAAUCGAGUGCAGUCUC--AAAACAUUUCUUAGAACUCGA---AACAAACAUAUAUAU- .(((.(.((((....--)))).))))(.((((.((((((.(((..---...))).))))))((...--...)).........)))).).---...............- ( -19.40, z-score = -2.06, R) >droAna3.scaffold_13337 7199665 102 - 23293914 AAGGGGGGAUUUCCAUCUCCCCUCUAAUAGUUGUGCCCCGUA-AC--AAAAAUCGGGUGCUGUCGC---AAACAAUUCUUAGAACUCAAUACUACAAACAUAUAUAUU ..((((((((....))))))))(((((.((((((..((((..-..--......))))(((....))---).)))))).)))))......................... ( -28.10, z-score = -3.30, R) >consensus AUGGCGCGGAAUUUA__UUCCACCCACUAGUUAUGCACUGGAUAC__AAAAAUCGAGUGCAAUCUC_AAAAACAUUACUUAGAACUCGA___AACAAACAUAUAUA__ .(((.(.((((......)))).)))).......((((((.(((........))).))))))............................................... (-12.52 = -13.63 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:47 2011