| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,067,288 – 17,067,402 |
| Length | 114 |
| Max. P | 0.960062 |

| Location | 17,067,288 – 17,067,402 |
|---|---|
| Length | 114 |
| Sequences | 14 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 74.49 |
| Shannon entropy | 0.55580 |
| G+C content | 0.52907 |
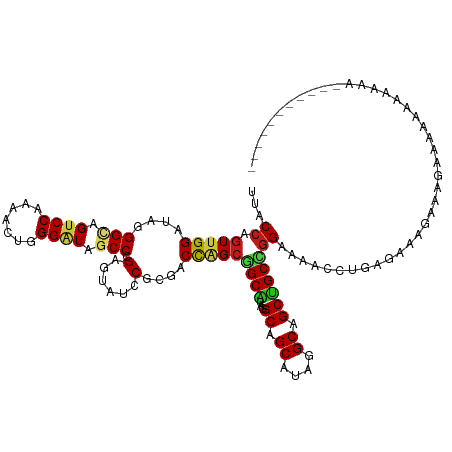
| Mean single sequence MFE | -36.31 |
| Consensus MFE | -20.26 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.960062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
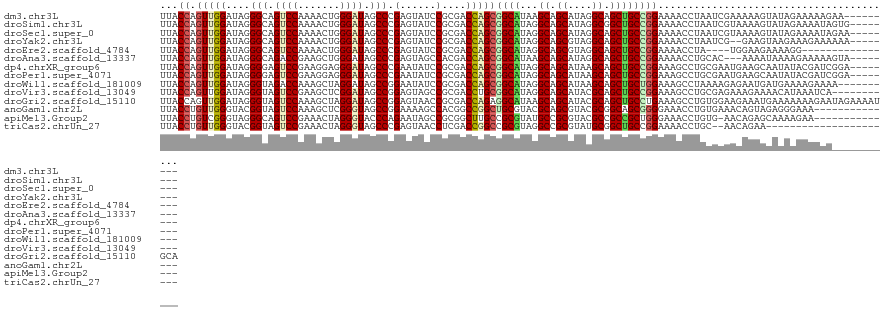
>dm3.chr3L 17067288 114 + 24543557 UUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAAGCAGCAUAGGCAGCUGCCGGAAAACCUAAUCGAAAAAGUAUAGAAAAAGAA--------- .(((...((((.((((((.((((.......)))).))(((..(((((((.....)))).))).(((((.......))))))))....)))).))))....)))...........--------- ( -32.30, z-score = -1.77, R) >droSim1.chr3L 16410306 115 + 22553184 UUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAGGCAGCAUAGGCGGCUGCCGGAAAACCUAAUCGUAAAAGUAUAGAAAAUAGUG-------- ((((..(((((...((((.((((.......)))).))))...(((((((.....)))).)))((((((.......)))))).......))))).)))).................-------- ( -35.80, z-score = -1.57, R) >droSec1.super_0 9127573 115 + 21120651 UUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAGGCAGCAUAGGCAGCUGCCGGAAAACCUAAUCGUAAAAGUAUAGAAAAUAGAA-------- ((((..(((((...((((.((((.......)))).))))...(((((((.....)))).)))((((((.......)))))).......))))).)))).................-------- ( -35.90, z-score = -2.27, R) >droYak2.chr3L 7193963 113 - 24197627 UUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAGGCAGCGUAGGCAGCUGCCGGAAAACCUAAUCG--GAAGUAAGAAAGAAAAAA-------- ...((.(((((...((((.((((.......)))).))))...(((((((.....)))).)))((((((.......)))))).......))))).)--).................-------- ( -38.70, z-score = -2.78, R) >droEre2.scaffold_4784 8986944 103 - 25762168 UUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAGGCAGCGUAGGCAGCUGCCGGAAAACCUA----UGGAAGAAAAGG---------------- ...(((...((...((((.((((.......)))).))))...(((((((.....)))).)))((((((.......))))))......))..----))).........---------------- ( -38.80, z-score = -2.69, R) >droAna3.scaffold_13337 7199207 112 + 23293914 UUACCAGUUGGAUAGGGCAGACCGAAGCUGGGAUAGCCCGAGUAGCCACGACCAGCGGCAUAAGCAGCAUAGGCAGCUGCCGGAAAACCUGCAC---AAAAUAAAAGAAAAAGUA-------- .......(((..((((.....(((..((((((....))).))).(((.(.....).)))....(((((.......))))))))....))))..)---))................-------- ( -29.20, z-score = -0.55, R) >dp4.chrXR_group6 13200003 115 + 13314419 UUACCAGUUGGAUAGGGGAGUCCGAAGGAGGGAUAGCCCGAAUAUCCGCGACCAGCGGCAUAGGCAGCAUAAGCAGCUGCCGGAAAGCCUGCGAAUGAAGCAAUAUACGAUCGGA-------- ...((.((((.(((.(((..(((......(((....)))...(((((((.....)))).)))((((((.......)))))))))...)))((.......))..))).)))).)).-------- ( -37.80, z-score = -1.37, R) >droPer1.super_4071 4304 115 + 4599 UUACCAGUUGGAUAGGGGAGUCCGAAGGAGGGAUAGCCCGAAUAUCCGCGACCAGCGGCAUAGGCAGCAUAAGCAGCUGCCGGAAAGCCUGCGAAUGAAGCAAUAUACGAUCGGA-------- ...((.((((.(((.(((..(((......(((....)))...(((((((.....)))).)))((((((.......)))))))))...)))((.......))..))).)))).)).-------- ( -37.80, z-score = -1.37, R) >droWil1.scaffold_181009 252304 113 - 3585778 UUACCAGUUGGAUAGGGUAGACCAAAGCUAGGAUAGCCGGAAUAUCCGCGACCAGCGGCAUAGGCAGCAUAAGCAGCUGCUGGAAAGCCUAAAAGAGAAUGAUGAAAAGAAAA---------- ......(((((...(((((..((...((((...)))).))..)))))....)))))(((...((((((.......)))))).....)))........................---------- ( -30.60, z-score = -1.70, R) >droVir3.scaffold_13049 20191577 112 - 25233164 UUACCAGUUGGAUAGGGUAGUCCGAAGCUCGGAUAGCCGGAGUAGCCGCGACCUGCGGCAUAGGCAGCAUACGCAGCUGCCGGAAAGCCUGCGAGAAAGAAAACAUAAAUCA----------- ...((....))...((((..(((...((((((....)).)))).(((((.....)))))...((((((.......)))))))))..))))......................----------- ( -37.50, z-score = -1.33, R) >droGri2.scaffold_15110 9909871 123 + 24565398 UUACCAGUUGGAUAGGGUAGUCCAAAGCUAGGAUAGCCGGAGUAACCGCGACCAGAGGCAUAAGCAGCAUACGCAGCUGCCUGAAAGCCUGUGGAAGAAAUGAAAAAAAGAAUAGAAAAUGCA ...((((((((((......)))))..))).))...(((((.....)))...(((.((((.((.(((((.......))))).))...)))).)))..........................)). ( -31.00, z-score = -1.32, R) >anoGam1.chr2L 35271953 109 + 48795086 UUACCUGUUGGGUACGGUAGUCCAAAGCUCGGGUAGCCGGAAAAGCCACGGCCGGCUGCGUACGCAGCGUACGCGGCAGCGGGGAAACCUGUGAAACAGUAGAGGGAAA-------------- ...(((((((.((((((....))...(((((.((((((((...........))))))))...)).))))))).)))).(((((....)))))..........)))....-------------- ( -44.40, z-score = -1.64, R) >apiMel3.Group2 9133414 108 + 14465785 UUACCUGUCGGGUAGGGCAGUCCGAAACUAGGGUACCCAGAAUAGCCGCGGCUUGCCGCGUAUGCCGCGUACGCCGCCGCUGGGAAACCUGUG-AACAGAGCAAAAGAA-------------- .(((((.(((((........))))).....)))))((((.....((.(((((.(((.(((.....)))))).))))).))))))....(((..-..)))..........-------------- ( -37.20, z-score = 0.12, R) >triCas2.chrUn_27 157326 99 - 823867 UUACCUGUUGGGUACGGUAGUCCGAAACUAGGGUAGCCCGAGUAACCUCGACCGGCCGCGUAGGCCGCGUAUGCGGCUGCCGGAAAACCUGC--AACAGAA---------------------- ....(((((((((.(((....))).......(((((((((((....))))(((((((.....))))).))....))))))).....)))..)--)))))..---------------------- ( -41.40, z-score = -1.88, R) >consensus UUACCAGUUGGAUAGGGCAGUCCAAAACUGGGAUAGCCCGAGUAUCCGCGACCAGCGGCAUAGGCAGCAUAGGCAGCUGCCGGAAAACCUGAGAAAGAAAGAAAAAAAAAAA___________ ...((.(((((....(((.((((.......)))).))).(......)....)))))((((...((.((....)).))))))))........................................ (-20.26 = -20.32 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:46 2011