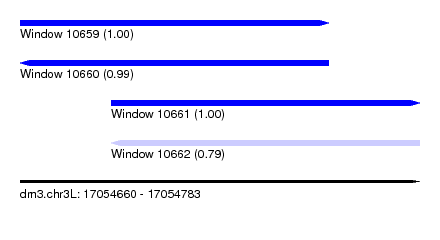
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,054,660 – 17,054,783 |
| Length | 123 |
| Max. P | 0.996727 |

| Location | 17,054,660 – 17,054,755 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.35 |
| Shannon entropy | 0.60805 |
| G+C content | 0.42479 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -14.76 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
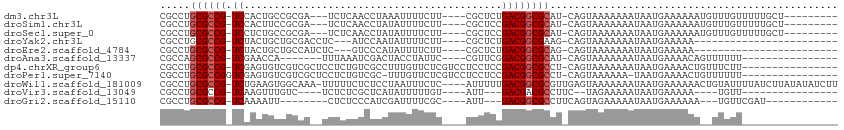
>dm3.chr3L 17054660 95 + 24543557 ----------UAAUAUUUU--GUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCCACUGCCGCGA-UCUCAACCUA-AAUUUUCUUCGCUCUGACGGCGCAUC-AGUAAAA ----------.........--......-.........(((((...(((((((((.......((((-..........-........))))...)))))))))..-.))))). ( -23.47, z-score = -2.40, R) >droSim1.chr3L 16396678 95 + 22553184 ----------UAAUAUUUU--GUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCCACUUCCGCGA-UCUCAACCUA-UAUUUUCUUCGCUCCGACGGCGCAUC-AGUAAAA ----------.........--......-.........(((((...(((((((((.......((((-..........-........))))...)))))))))..-.))))). ( -23.47, z-score = -3.10, R) >droSec1.super_0 9115069 95 + 21120651 ----------UAAUAUUUU--GUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCCUCUGCCGCGA-UCUCAACCUA-UAUUUUCUUCGCUCCGACGGCGCAUC-AGUAAAA ----------.........--......-.........(((((...(((((((((.......((((-..........-........))))...)))))))))..-.))))). ( -23.47, z-score = -2.55, R) >droYak2.chr3L 7180454 97 - 24197627 ----------UAAUAUUUUUUGUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCUACUGCUGCGA-CCUCAUCCAA-UAUUUUCUUCGCUCUGACGGCGAAGC-AGUAAAA ----------.................-.........(((((((...(((((((.......((((-..........-........))))...)))))))..))-.))))). ( -20.87, z-score = -0.96, R) >droEre2.scaffold_4784 8974567 95 - 25762168 ----------UAAUAUUUU--GUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCUACUGCUGCCA-UCUCGUCCCA-UAUUUUCUUCGCUCUGACGGCGCAGC-AGUAAAA ----------.........--......-.........(((((..((((((((((....((.....-..........-..........))...)))))))))).-.))))). ( -24.41, z-score = -2.37, R) >droAna3.scaffold_13337 7187273 91 + 23293914 ----------UCUUAUUUU--GUUCAA-UUUAAAGCUUUUGCGCCAGCGCCGUCGA-----ACCA-UUUAAAUCGACUACCUAUUCCGUUCGGACGGCGCAUC-AGUAAAA ----------.........--......-......(((..((((((......(((((-----....-......))))).......(((....))).))))))..-))).... ( -22.40, z-score = -2.21, R) >droWil1.scaffold_181009 232990 109 - 3585778 AUAUAUUUUUUAGGGUUUUU-GUUUUG-GUUAAAGCUUUUGCGCCUGCGCCGUCUGAAGUGGCAAAUUUUUCUCUCCUAAUUUCUCAUUUUUGACGGCGCGUUGAGUAAAA ...(((((...(((((....-(((((.-...)))))....)).)))((((((((.(((((((.(((((..........))))).))))))).))))))))...)))))... ( -30.40, z-score = -2.77, R) >droVir3.scaffold_13049 20162956 89 - 25233164 -----------UUUAUUAU--GUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCAAGUUUGUCUC-UCUCGCUCAU-----AUUUUUGUAUUGACGACGCCUUC--UAGAA -----------........--.(((..-......((....))....(((.((((((.........-..........-----.........)))))).)))....--..))) ( -11.69, z-score = 0.80, R) >droMoj3.scaffold_6680 21313527 80 - 24764193 ----------UUAUAUUAU--UCUCAAAUUUAAAGCUUUUGCGCCUGCGCCGUCAAGUUU------UCCUGUUCAC-----GUU--------GACGACGCCUUCAGUAGAA ----------.........--................(((((....(((.((((((((..------........))-----.))--------)))).))).....))))). ( -13.60, z-score = -0.40, R) >droGri2.scaffold_15110 9896315 87 + 24565398 -----------UUAAUUAU--GUUCAA-UUUAAAGCUUUUGCGCCUGCGCCGUCAAAAUU--CUC-UCCCAUCGAU-----UUUC--GCAUUGACGGCGCCUUCAGUAGAA -----------........--......-.........(((((....((((((((((....--...-..........-----....--...)))))))))).....))))). ( -19.46, z-score = -1.57, R) >consensus __________UAAUAUUUU__GUUCAA_UUUAAAGCUUUUGCGCCUGCGCCGUCAACUGC_GCGA_UCUCAACCAA_UAUUUUCUUCGCUCUGACGGCGCAUC_AGUAAAA .....................................(((((....((((((((......................................)))))))).....))))). (-14.76 = -14.95 + 0.19)
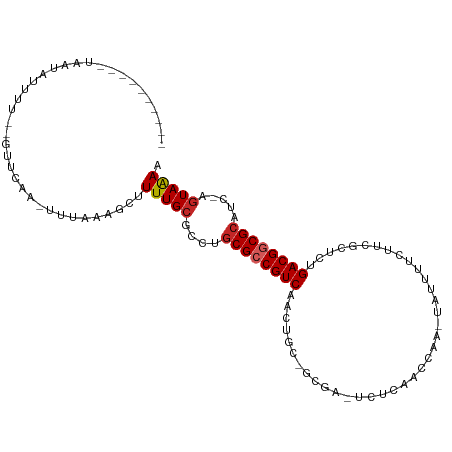
| Location | 17,054,660 – 17,054,755 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.35 |
| Shannon entropy | 0.60805 |
| G+C content | 0.42479 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17054660 95 - 24543557 UUUUACU-GAUGCGCCGUCAGAGCGAAGAAAAUU-UAGGUUGAGA-UCGCGGCAGUGGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAAC--AAAAUAUUA---------- .......-..(((((((((...((((....(((.-...)))....-)))).......)))))))))(((......))).....-......--.........---------- ( -25.00, z-score = -0.77, R) >droSim1.chr3L 16396678 95 - 22553184 UUUUACU-GAUGCGCCGUCGGAGCGAAGAAAAUA-UAGGUUGAGA-UCGCGGAAGUGGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAAC--AAAAUAUUA---------- .......-..(((((((((.(.((((....(((.-...)))....-)))).....).)))))))))(((......))).....-......--.........---------- ( -25.80, z-score = -1.39, R) >droSec1.super_0 9115069 95 - 21120651 UUUUACU-GAUGCGCCGUCGGAGCGAAGAAAAUA-UAGGUUGAGA-UCGCGGCAGAGGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAAC--AAAAUAUUA---------- .......-..(((((((((.(.((((....(((.-...)))....-))))..)....)))))))))(((......))).....-......--.........---------- ( -24.70, z-score = -0.88, R) >droYak2.chr3L 7180454 97 + 24197627 UUUUACU-GCUUCGCCGUCAGAGCGAAGAAAAUA-UUGGAUGAGG-UCGCAGCAGUAGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAACAAAAAAUAUUA---------- .((((((-(((.((((.(((...(.((.......-)).).)))))-.)).)))))))))..(((....)))............-.................---------- ( -22.50, z-score = -0.13, R) >droEre2.scaffold_4784 8974567 95 + 25762168 UUUUACU-GCUGCGCCGUCAGAGCGAAGAAAAUA-UGGGACGAGA-UGGCAGCAGUAGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAAC--AAAAUAUUA---------- .((((((-(((((.(((((...............-...)))).).-..)))))))))))..(((....)))............-......--.........---------- ( -24.27, z-score = -0.77, R) >droAna3.scaffold_13337 7187273 91 - 23293914 UUUUACU-GAUGCGCCGUCCGAACGGAAUAGGUAGUCGAUUUAAA-UGGU-----UCGACGGCGCUGGCGCAAAAGCUUUAAA-UUGAAC--AAAAUAAGA---------- .......-...((((((((.((((.(..(((((.....)))))..-).))-----)))))))))).(((......))).....-......--.........---------- ( -23.90, z-score = -1.03, R) >droWil1.scaffold_181009 232990 109 + 3585778 UUUUACUCAACGCGCCGUCAAAAAUGAGAAAUUAGGAGAGAAAAAUUUGCCACUUCAGACGGCGCAGGCGCAAAAGCUUUAAC-CAAAAC-AAAAACCCUAAAAAAUAUAU ...........((((((((.....(((....)))(((((......))).))......))))))))((((......))))....-......-.................... ( -23.40, z-score = -2.66, R) >droVir3.scaffold_13049 20162956 89 + 25233164 UUCUA--GAAGGCGUCGUCAAUACAAAAAU-----AUGAGCGAGA-GAGACAAACUUGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAAC--AUAAUAAA----------- .....--....((((((((((..((.....-----.))..(....-)........))))))))))((((......))))....-......--........----------- ( -18.40, z-score = -1.26, R) >droMoj3.scaffold_6680 21313527 80 + 24764193 UUCUACUGAAGGCGUCGUC--------AAC-----GUGAACAGGA------AAACUUGACGGCGCAGGCGCAAAAGCUUUAAAUUUGAGA--AUAAUAUAA---------- .....((.(((((((((((--------((.-----((........------..))))))))))))((((......))))....))).)).--.........---------- ( -21.30, z-score = -2.21, R) >droGri2.scaffold_15110 9896315 87 - 24565398 UUCUACUGAAGGCGCCGUCAAUGC--GAAA-----AUCGAUGGGA-GAG--AAUUUUGACGGCGCAGGCGCAAAAGCUUUAAA-UUGAAC--AUAAUUAA----------- ...........((((((((((...--....-----.((.....))-...--....))))))))))((((......))))....-......--........----------- ( -22.36, z-score = -1.30, R) >consensus UUUUACU_GAUGCGCCGUCAGAGCGAAGAAAAUA_UAGGAUGAGA_UCGC_GCAGUAGACGGCGCAGGCGCAAAAGCUUUAAA_UUGAAC__AAAAUAUUA__________ ...........((((((((......................................))))))))((((......))))................................ (-16.16 = -16.20 + 0.04)

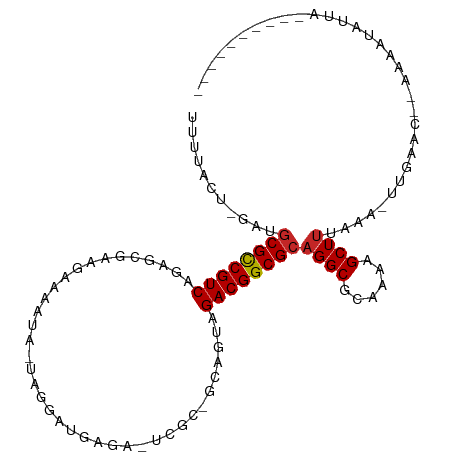
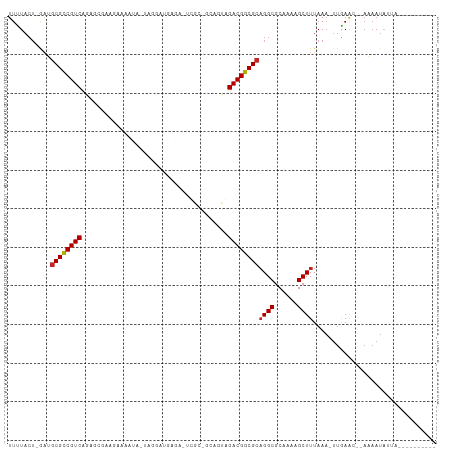
| Location | 17,054,688 – 17,054,783 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.60 |
| Shannon entropy | 0.72326 |
| G+C content | 0.45219 |
| Mean single sequence MFE | -20.86 |
| Consensus MFE | -9.18 |
| Energy contribution | -9.36 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.996210 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 17054688 95 + 24543557 CGCCUGCGCCG-UCCACUGCCGCGA---UCUCAACCUAAAUUUUCUU----CGCUCUGACGGCGCAU-CAGUAAAAAAAUAAUGAAAAAAUGUUUGUUUUUGCU--------- ....(((((((-((.......((((---..................)----)))...))))))))).-.((((((((...(((........)))..))))))))--------- ( -23.97, z-score = -2.60, R) >droSim1.chr3L 16396706 95 + 22553184 CGCCUGCGCCG-UCCACUUCCGCGA---UCUCAACCUAUAUUUUCUU----CGCUCCGACGGCGCAU-CAGUAAAAAAAUAAUGAAAAAAUGUUUGUUUUUGCU--------- ....(((((((-((.......((((---..................)----)))...))))))))).-.((((((((...(((........)))..))))))))--------- ( -23.97, z-score = -3.01, R) >droSec1.super_0 9115097 95 + 21120651 CGCCUGCGCCG-UCCUCUGCCGCGA---UCUCAACCUAUAUUUUCUU----CGCUCCGACGGCGCAU-CAGUAAAAAAAUAAUGAAAAAAUGUUUGUUUUUGCU--------- ....(((((((-((.......((((---..................)----)))...))))))))).-.((((((((...(((........)))..))))))))--------- ( -23.97, z-score = -2.69, R) >droYak2.chr3L 7180484 80 - 24197627 CGCCUGCGCCG-UCUACUGCUGCGACCUC---AUCCAAUAUUUUCUU----CGCUCUGACGGCGAAG-CAGUAAAAAAAUAAUGAAAAA------------------------ .((...(((((-((.......((((....---..............)----)))...)))))))..)-)....................------------------------ ( -18.47, z-score = -1.79, R) >droEre2.scaffold_4784 8974595 80 - 25762168 CGCCUGCGCCG-UCUACUGCUGCCAUCUC---GUCCCAUAUUUUCUU----CGCUCUGACGGCGCAG-CAGUAAAAAAAUAAUGAAAAA------------------------ .((.(((((((-((....((.........---...............----.))...))))))))))-)....................------------------------ ( -21.31, z-score = -2.69, R) >droAna3.scaffold_13337 7187301 84 + 23293914 CGCCAGCGCCG-UCGAACCA-------UUUAAAUCGACUACCUAUUC----CGUUCGGACGGCGCAU-CAGUAAAAAAAUAAUGAAAACAGUUUUUU---------------- .....((((((-((((((..-------....................----.)))).))))))))..-.....(((((((..(....)..)))))))---------------- ( -19.50, z-score = -1.98, R) >dp4.chrXR_group6 13185876 95 + 13314419 CGCCUGCGCCG-UCGAGUGUCGUCGCUCCUCUGUCGCCUUUGUUCUCGUCCUCCUCCGACGGCGCCU-CAGUAAAAAAAUAAUGAAAACUGUUUCUU---------------- .....((((((-(((((((....)))))....(.((..........)).).......))))))))..-((((...............))))......---------------- ( -21.66, z-score = -1.99, R) >droPer1.super_7140 1415 94 + 1540 CGCCUGCGCCGGUCGAGUGUCGUCGCUCCUCUGUCGC-UUUGUUCUCGUCCUCCUCCGACGGCGCCU-CAGUAAAAAA-UAAUGAAAACUGUUUUUU---------------- ((((.(((.(((..(((((....)))))..))).)))-.........(((.......)))))))...-....((((((-((.(....).))))))))---------------- ( -24.70, z-score = -2.16, R) >droWil1.scaffold_181009 233029 107 - 3585778 CGCCUGCGCCG-UCUGAAGUGGCAAA-UUUUUCUCUCCUAAUUUCUC----AUUUUUGACGGCGCGUUGAGUAAAAAAAUAAUGAAAAAACUGUAUUUUAUCUUAUAUAUCUU .....((((((-((.(((((((.(((-((..........))))).))----))))).))))))))..((((....((((((..(......)..))))))..))))........ ( -25.00, z-score = -3.27, R) >droVir3.scaffold_13049 20162983 79 - 25233164 CGCCUGCGCCG-UCAAGUUUGUC----UCUCUCGCUCAUAUUUUUGU----AUU---GACGACGCCUUC--UAGAAAAAUAAUGAAAAA----UGUU---------------- .....(((.((-((((.......----....................----.))---)))).)))....--..................----....---------------- ( -9.69, z-score = 0.78, R) >droGri2.scaffold_15110 9896342 82 + 24565398 CGCCUGCGCCG-UCAAAAUU--------CUCUCCCAUCGAUUUUCGC----AUU---GACGGCGCCUUCAGUAGAAAAAUAAUGAAAAAA---UGUUCGAU------------ .....((((((-((((....--------...................----.))---)))))))).((((.((......)).))))....---........------------ ( -17.26, z-score = -1.02, R) >consensus CGCCUGCGCCG_UCCACUGCCGCGA__UCUCUAUCCCAUAUUUUCUU____CGCUCCGACGGCGCAU_CAGUAAAAAAAUAAUGAAAAAA__UUUUU________________ .....((((((.((...........................................))))))))................................................ ( -9.18 = -9.36 + 0.18)
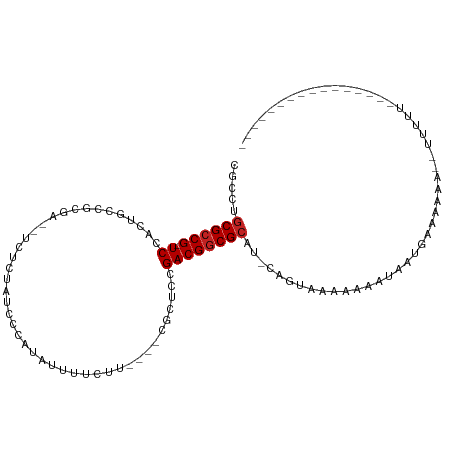
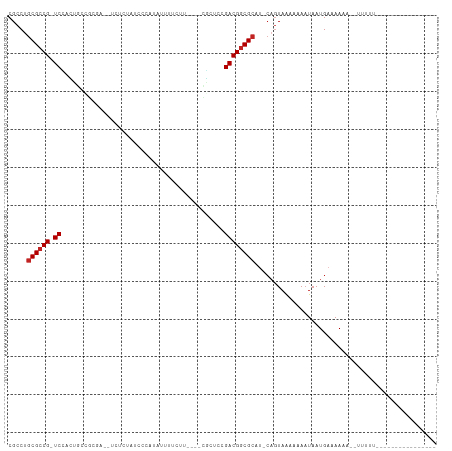
| Location | 17,054,688 – 17,054,783 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 62.60 |
| Shannon entropy | 0.72326 |
| G+C content | 0.45219 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -9.82 |
| Energy contribution | -9.83 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792339 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 17054688 95 - 24543557 ---------AGCAAAAACAAACAUUUUUUCAUUAUUUUUUUACUG-AUGCGCCGUCAGAGCG----AAGAAAAUUUAGGUUGAGA---UCGCGGCAGUGGA-CGGCGCAGGCG ---------.((................(((.((......)).))-)(((((((((...(((----(....(((....)))....---)))).......))-))))))).)). ( -25.30, z-score = -1.15, R) >droSim1.chr3L 16396706 95 - 22553184 ---------AGCAAAAACAAACAUUUUUUCAUUAUUUUUUUACUG-AUGCGCCGUCGGAGCG----AAGAAAAUAUAGGUUGAGA---UCGCGGAAGUGGA-CGGCGCAGGCG ---------.((................(((.((......)).))-)(((((((((.(.(((----(....(((....)))....---)))).....).))-))))))).)). ( -26.10, z-score = -1.54, R) >droSec1.super_0 9115097 95 - 21120651 ---------AGCAAAAACAAACAUUUUUUCAUUAUUUUUUUACUG-AUGCGCCGUCGGAGCG----AAGAAAAUAUAGGUUGAGA---UCGCGGCAGAGGA-CGGCGCAGGCG ---------.((................(((.((......)).))-)(((((((((.(.(((----(....(((....)))....---))))..)....))-))))))).)). ( -25.00, z-score = -1.23, R) >droYak2.chr3L 7180484 80 + 24197627 ------------------------UUUUUCAUUAUUUUUUUACUG-CUUCGCCGUCAGAGCG----AAGAAAAUAUUGGAU---GAGGUCGCAGCAGUAGA-CGGCGCAGGCG ------------------------....................(-((((((((((...(((----(.....((.....))---....)))).......))-))))).)))). ( -20.20, z-score = -0.47, R) >droEre2.scaffold_4784 8974595 80 + 25762168 ------------------------UUUUUCAUUAUUUUUUUACUG-CUGCGCCGUCAGAGCG----AAGAAAAUAUGGGAC---GAGAUGGCAGCAGUAGA-CGGCGCAGGCG ------------------------..............(((((((-((((.(((((......----............)))---).)...)))))))))))-..((....)). ( -21.07, z-score = -0.58, R) >droAna3.scaffold_13337 7187301 84 - 23293914 ----------------AAAAAACUGUUUUCAUUAUUUUUUUACUG-AUGCGCCGUCCGAACG----GAAUAGGUAGUCGAUUUAAA-------UGGUUCGA-CGGCGCUGGCG ----------------............(((.((......)).))-).((((((((.((((.----(..(((((.....)))))..-------).))))))-))))))..... ( -21.90, z-score = -0.99, R) >dp4.chrXR_group6 13185876 95 - 13314419 ----------------AAGAAACAGUUUUCAUUAUUUUUUUACUG-AGGCGCCGUCGGAGGAGGACGAGAACAAAGGCGACAGAGGAGCGACGACACUCGA-CGGCGCAGGCG ----------------......((((...............))))-..((((((((((.(..(..((........(....).......)).)..).).)))-))))))..... ( -23.62, z-score = -0.41, R) >droPer1.super_7140 1415 94 - 1540 ----------------AAAAAACAGUUUUCAUUA-UUUUUUACUG-AGGCGCCGUCGGAGGAGGACGAGAACAAA-GCGACAGAGGAGCGACGACACUCGACCGGCGCAGGCG ----------------......((((........-......))))-..(((((.((....))((.((((......-((..(...)..)).......)))).)))))))..... ( -20.96, z-score = 0.54, R) >droWil1.scaffold_181009 233029 107 + 3585778 AAGAUAUAUAAGAUAAAAUACAGUUUUUUCAUUAUUUUUUUACUCAACGCGCCGUCAAAAAU----GAGAAAUUAGGAGAGAAAAA-UUUGCCACUUCAGA-CGGCGCAGGCG ((((...((((((.(((((...))))).)).))))..)))).......((((((((.....(----((....)))(((((......-))).))......))-))))))..... ( -21.70, z-score = -1.85, R) >droVir3.scaffold_13049 20162983 79 + 25233164 ----------------AACA----UUUUUCAUUAUUUUUCUA--GAAGGCGUCGUC---AAU----ACAAAAAUAUGAGCGAGAGA----GACAAACUUGA-CGGCGCAGGCG ----------------....----..................--(...((((((((---((.----.((......))..(....).----.......))))-))))))...). ( -16.00, z-score = -0.98, R) >droGri2.scaffold_15110 9896342 82 - 24565398 ------------AUCGAACA---UUUUUUCAUUAUUUUUCUACUGAAGGCGCCGUC---AAU----GCGAAAAUCGAUGGGAGAG--------AAUUUUGA-CGGCGCAGGCG ------------........---...(((((.((......)).)))))((((((((---((.----.......((.....))...--------....))))-))))))..... ( -20.56, z-score = -0.73, R) >consensus ________________AAAAA__UUUUUUCAUUAUUUUUUUACUG_AUGCGCCGUCAGAGCG____AAGAAAAUAUGGGAUAAAGA__UCGCGACAGUAGA_CGGCGCAGGCG ................................................((((((((...........................................)).))))))..... ( -9.82 = -9.83 + 0.01)
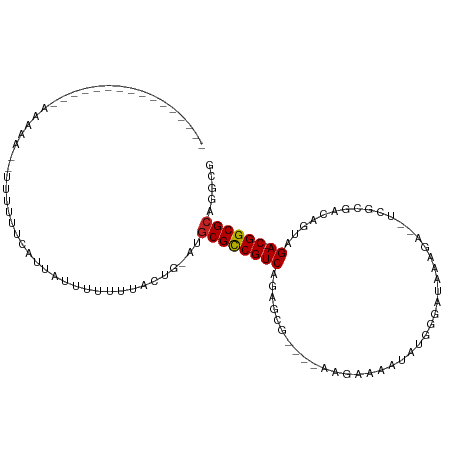
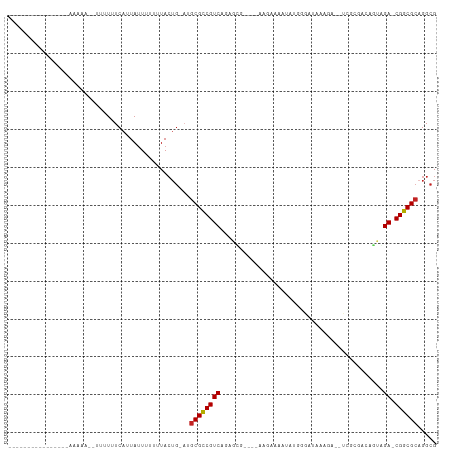
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:42 2011