| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,039,592 – 17,039,690 |
| Length | 98 |
| Max. P | 0.866081 |

| Location | 17,039,592 – 17,039,690 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.47 |
| Shannon entropy | 0.51337 |
| G+C content | 0.45175 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17039592 98 + 24543557 -------------AAGGAGUACUUAUUGUUUUUAC-AACU-GCAGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUAAACG -------------..(...((((((((((((((((-.(((-.(.((((((..((((.....)))).(((....))).))))))...).))).).)))))))))))))))..). ( -31.50, z-score = -2.65, R) >droSim1.chr3L 16380688 99 + 22553184 -------------AAGGAGUACUUAUUGUUUUUGGCAACU-GCAGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUAAACG -------------..(...((((((((((((((.((....-))..(((.(((((((((.....((.(((....))).))..)))))).))).)))))))))))))))))..). ( -33.30, z-score = -2.81, R) >droSec1.super_0 9100095 99 + 21120651 -------------AUGGAGUACUUAUUGUUUUUGGCAACU-GCAGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAAGUCGAUGAGGACAGUAAGUAAACG -------------..(...(((((((((((((((((..((-...((((((..((((.....)))).(((....))).))))))..))..)))))...))))))))))))..). ( -31.70, z-score = -2.50, R) >droYak2.chr3L 7163762 96 - 24197627 -------------UGGGAGUACUUAU-GCUUUUGG-AACU-GCAGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUAUAU- -------------.....((((((((-.(((((.(-(.((-.(.((((((..((((.....)))).(((....))).))))))..).)).)).).))))...))))))))..- ( -26.70, z-score = -0.74, R) >droEre2.scaffold_4784 8959053 97 - 25762168 -------------UGGCUGUACUUAU-GCUUUUGG-AACU-GCAGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUAAACG -------------..(((((.(((.(-((......-....-))).(((.(((((((((.....((.(((....))).))..)))))).))).)))))).)))))......... ( -26.50, z-score = -0.42, R) >droAna3.scaffold_13337 7171949 97 + 23293914 ----------AACACGGAAUAAUAGUUC------UUAAUAUCCAGGUCAGCCCUUCGACAAGAAGAUCUUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUAAAAG ----------..............((((------(((...(((.((((((..((((..((((.....))))))))..))))))..)))......)))))))............ ( -24.30, z-score = -1.21, R) >dp4.chrXR_group6 13172745 112 + 13314419 AUGUCACCUUAACACCUAACAUUGUCCCAUUUGAUUAAUUUGUAGGUCAGCCCUUCGACAAGAAGAUCUUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUGAUC- ..(((((.(((.....))).(((((((((((.(((...(((...((((((..((((..((((.....))))))))..))))))..))).))))))).)))))))..))))).- ( -29.90, z-score = -0.67, R) >droPer1.super_88 211165 112 + 213009 AUGUCACCUUAACACCUAACAUUGUCCCAUUUGAUUAAUUUGUAGGUCAGCCCUUCGACAAGAAGAUCUUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUGAUC- ..(((((.(((.....))).(((((((((((.(((...(((...((((((..((((..((((.....))))))))..))))))..))).))))))).)))))))..))))).- ( -29.90, z-score = -0.67, R) >droVir3.scaffold_13049 20150585 94 - 25233164 ----------------CUGUAAUCAUAAAUUGAUCGCAUU--UAGGUCAGCCCUUCGACAAGAAGAUCUUGGAGGAACUGAUCGAGGAGGUUGACGAAGACAGUAAGUACAC- ----------------.((..((((.....))))..))..--...(((((((((((((.....((.(((....))).))..)))))).))))))).................- ( -25.60, z-score = -1.94, R) >droMoj3.scaffold_6680 21300849 106 - 24764193 -------CUGGAACUUUUAUUUUCAUUGGUCUUACCACUUUAUAGGUCAGCCCUUCGACAAGAAGAUUCUGGAGGAACUGAUCGAAGAGGUCGAUGAGGACAGUAAGUAUUUU -------.....((((...(((((((((..((.....((((...((((((.((((((............))))))..))))))))))))..)))))))))....))))..... ( -25.90, z-score = -0.21, R) >droGri2.scaffold_15110 9883732 94 + 24565398 -----------------AAACUACUUUGGUUUUGUCUCUUGA-AGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAGGUUGAUGAGGACAGUAAGUAUAC- -----------------....(((((.....(((((.(((..-..(((((((((((((.....((.(((....))).))..)))))).))))))))))))))).)))))...- ( -28.80, z-score = -1.44, R) >consensus _____________ACGGAGUACUUAUUGAUUUUGGCAAUU_GCAGGUCAGCCCUUCGACAAGAAGAUCCUGGAGGAACUGAUCGAGGAGGUCGAUGAGGACAGUAAGUAAAC_ .............................................(((.(((((((((.....((.(((....))).))..)))))).))).))).................. (-16.19 = -15.80 + -0.39)


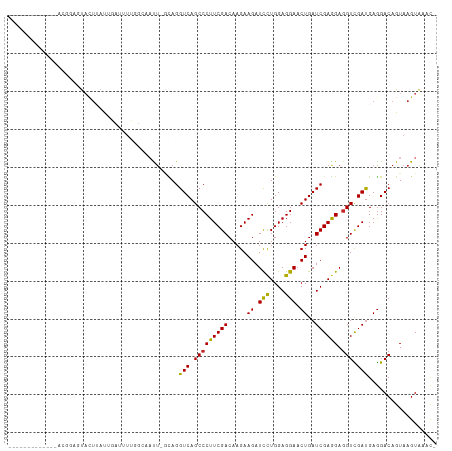
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:37 2011