
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,036,966 – 17,037,067 |
| Length | 101 |
| Max. P | 0.633148 |

| Location | 17,036,966 – 17,037,067 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 63.94 |
| Shannon entropy | 0.75440 |
| G+C content | 0.39873 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.13 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.35 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17036966 101 + 24543557 AUUAUGCUGGUUACCGGCAACUUCUUCAGCGGCUUUGCAUUCUGUUUGGAGGUGGGUAAC--AUUUG--AGGUUUAUGAUGUAAGAGAUCUAAAGUGUAUUAUCC- ....((((((...))))))((((((.(((..((...))...)))...))))))(((((((--((((.--((((((.(......).)))))).))))))..)))))- ( -26.20, z-score = -1.14, R) >droAna3.scaffold_13337 7169454 93 + 23293914 AUCAUGUUGGUGACAGGAAACUUCUUCAGUGGUUUCGCAUUCUGCUUGGAGGUGAGUAAC--AA-----UACAAGAAAACUUCAGGGAAUUUUAUUCAAA------ .((((((((....)).((((((........))))))))))....(((((((.(..(((..--..-----)))..)....)))))))))............------ ( -15.40, z-score = 1.14, R) >droEre2.scaffold_4784 8956147 98 - 25762168 AUUAUGCUGGUUACCGGCAACUUCUUCAGCGGUUUUGCCUUCUGCUUGGAGGUGGGUUAU--AUAAA--AGGUUCUUCAUGUAAGAGAUCUAAAUUGUAUUC---- ....((((((...))))))((((((..(((((.........))))).)))))).....((--((((.--((((((((.....)))).))))...))))))..---- ( -21.30, z-score = -0.14, R) >droYak2.chr3L 7160894 98 - 24197627 AUUAUGCUGGUUACCGGCAACUUCUUCAGCGGCUUUGCCUUCUGCUUGGAGGUGCGUUAU--AGAAG--CGGUUUUUGAUGUAUGAGACUUAAAUUGUAUCC---- ....((((((...)))))).(((((..((((.(...((((((.....)))))))))))..--)))))--.((((((........))))))............---- ( -24.80, z-score = -0.40, R) >droSec1.super_0 9097501 98 + 21120651 AUUAUGCUGGUUACCGGCAACUUCUUCAGCGGCUUUGCCUUCUGCUUGGAGGUGGGUAAU--AUUUG--AGAUUUAUGAUGUAAGCGAUCUAAUUUGUAUCC---- ....((((((...))))))((((((.(((.(((...)))..)))...))))))(((((..--.....--(((((..(......)..)))))......)))))---- ( -22.82, z-score = -0.10, R) >droSim1.chr3L 16378084 98 + 22553184 AUUAUGCUGGUUACCGGCAACUUCUUCAGCGGCUUUGCCUUCUGCUUGGAGGUGGGUAAU--AUUUG--AGAUUUAUGAUGUGAGCGAUCUAAAUUGUAUCC---- ....((((((...))))))((((((.(((.(((...)))..)))...))))))(((((..--((((.--(((((............)))))))))..)))))---- ( -23.10, z-score = -0.02, R) >droWil1.scaffold_180955 240274 99 + 2875958 GUUAUGCUGAUCACAGGAAACUUCUUUAGUGGAUUGGCCUUUUGUCUGGAGGUAAAACUU--GUAAAUCUAUUUUGCCAAGUCGCAUCUAAUUAGUUUUAC----- ......(((....)))((((((...((((((((((.((((((.....)))))).......--(((((.....)))))..)))).)).))))..))))))..----- ( -18.60, z-score = 0.04, R) >droPer1.super_32 808356 104 + 992631 AUCAUGCUGGUCACGGGAAACUUUUUCAGUGGAUUCGCCUUCUGCUUGGAGGUCAGUAUA--AGGCGUCUUCACUACUACAUCAUAUACUAUUAUUAUAUCAAUCG ...(((.((((...((....)).....((((((..((((((.((((........)))).)--)))))..)))))))))))))........................ ( -23.20, z-score = -1.80, R) >dp4.chrXR_group8 8536981 104 - 9212921 AUCAUGCUGGUCACGGGAAACUUUUUCAGUGGAUUCGCCUUCUGCUUGGAGGUCAGUAUA--AGGCGUCUUCACUACUACAUCAUAUACUAUUAUUAUAUCAAUCG ...(((.((((...((....)).....((((((..((((((.((((........)))).)--)))))..)))))))))))))........................ ( -23.20, z-score = -1.80, R) >triCas2.ChLG3 24545947 100 + 32080666 GUGAUGGUGGCCACUGGCGACUUCACCGAUGGCUUGCAAUUUUUUUCUACUGUAGUUUAUUAAAUGAUCAUUUUUCCCAAAAGUUUGAUUUCAAUUAUAU------ ((((.(((.(((...))).))))))).(((((.(((((............))))).)))))....(((((..(((.....)))..)))))..........------ ( -15.50, z-score = 0.68, R) >consensus AUUAUGCUGGUUACCGGCAACUUCUUCAGCGGCUUUGCCUUCUGCUUGGAGGUGGGUAAU__AUUAG__AGCUUUAUGAUGUAAGAGAUUUAAAUUGUAUCC____ ....((((((.....(....)....)))))).....((((((.....))))))..................................................... (-10.43 = -10.13 + -0.30)

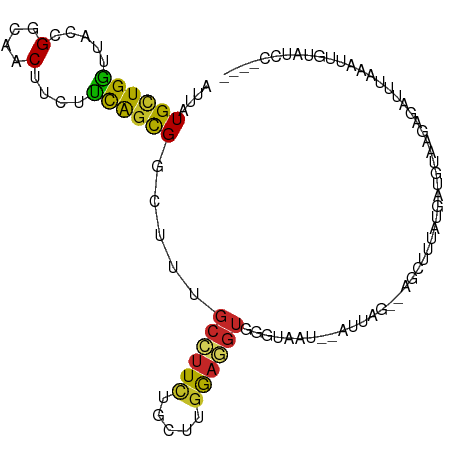
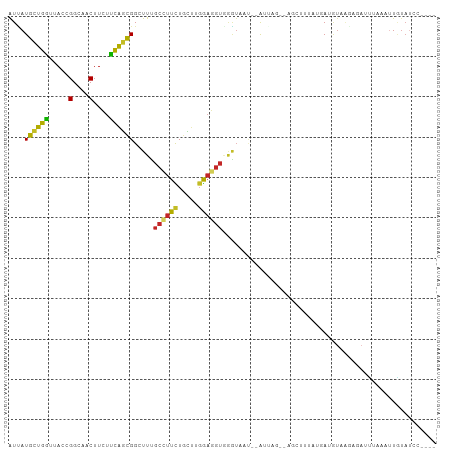
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:36 2011