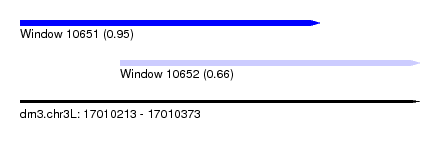
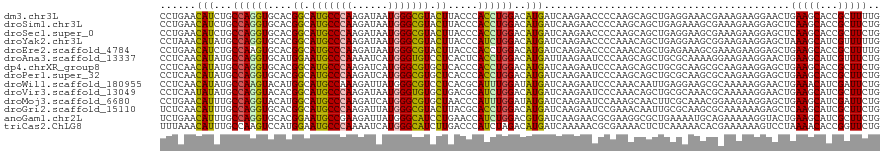
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 17,010,213 – 17,010,373 |
| Length | 160 |
| Max. P | 0.946562 |

| Location | 17,010,213 – 17,010,333 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.37335 |
| G+C content | 0.48722 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -23.28 |
| Energy contribution | -21.67 |
| Covariance contribution | -1.61 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
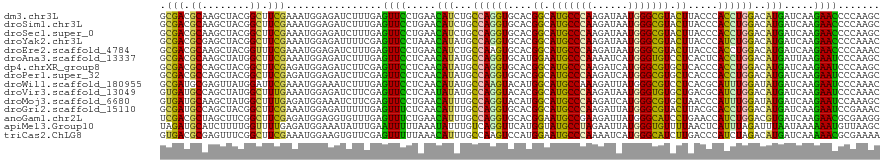
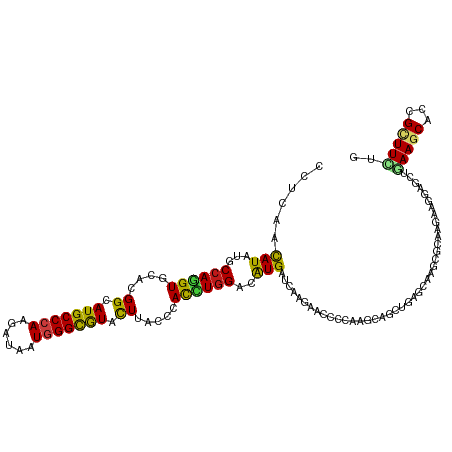
>dm3.chr3L 17010213 120 + 24543557 GCGACGCAAGCUACGGCUUCGAAAUGGAGAUCUUUGAGUUCCUGAACAUCUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGC ((..((.((((....))))))............(((.((((.(((.(((...(((((((...((.(((((((......))))))).))....)))))))..))).))).))))..))))) ( -41.80, z-score = -3.06, R) >droSim1.chr3L 16352307 120 + 22553184 GCGACGCAAGCUACGGCUUCGAAAUGGAGAUCUUUGAGUUCCUGAACAUCUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGC ((..((.((((....))))))............(((.((((.(((.(((...(((((((...((.(((((((......))))))).))....)))))))..))).))).))))..))))) ( -41.80, z-score = -3.06, R) >droSec1.super_0 9070808 120 + 21120651 GCGACGCAAGCUACGGCUUCGAAAUGGAGAUCUUUGAGUUCCUGAACAUCUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGC ((..((.((((....))))))............(((.((((.(((.(((...(((((((...((.(((((((......))))))).))....)))))))..))).))).))))..))))) ( -41.80, z-score = -3.06, R) >droYak2.chr3L 7133696 120 - 24197627 GCGACGCGAGCUACGGCUUCGAAAUGGAGAUUUUCGAGUUCCUAAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCAUCUGGACAUGAUCAAGAACCCCAAAC ....((.((((....))))))...(((.(.((((.(((((....))).(((((((((((...((.(((((((......))))))).))....))))))).)))).)).)))).))))... ( -35.60, z-score = -1.92, R) >droEre2.scaffold_4784 8929480 120 - 25762168 GCGACGCAAGCUACGGUUUCGAAAUGGAGAUCUUUGAGUUCCUGAACAUCUGCCAAGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAAC ((.......))...((((((......))))))((((.((((.(((.(((...(((.(((...((.(((((((......))))))).))....))).)))..))).))).))))..)))). ( -33.20, z-score = -1.43, R) >droAna3.scaffold_13337 9684617 120 - 23293914 GCGACGCAAGCUAUGGCUUCGAAAUGGAGAUCUUUGAGUUCCUCAACAUAUGCCAGGUGCAUGGAAUGCCCAAAAUCAUGGGUGUCCUCACUCACCUGGACAUGAUUAAGAAUCCCAAGC ((..((.((((....))))))...(((.((((((((((...))))...(((((((((((...(((.((((((......))))))))).....))))))).))))...))).)))))).)) ( -39.10, z-score = -2.21, R) >dp4.chrXR_group8 8513009 120 - 9212921 GCGACGCCAGCUACGGCUUCGAGAUGGAGAUCUUCGAGUUCCUCAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUCAUGGGCGUGCUCACCCACCUGGACAUGAUCAAGAAUCCCAAGC .(((.(((......))).)))...(((.((((((.(((((....))).(((((((((((...((((((((((......))))))))))....))))))).)))).))))).))))))... ( -48.10, z-score = -3.46, R) >droPer1.super_32 784389 120 + 992631 GCGACGCCAGCUACGGCUUCGAGAUGGAGAUCUUCGAGUUCCUCAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUCAUGGGCGUGCUCACCCACCUGGACAUGAUCAAGAAUCCCAAGC .(((.(((......))).)))...(((.((((((.(((((....))).(((((((((((...((((((((((......))))))))))....))))))).)))).))))).))))))... ( -48.10, z-score = -3.46, R) >droWil1.scaffold_180955 216216 120 + 2875958 GCGAUGCGAGUUAUGGAUUCGAAAUGGAAAUCUUUGAGUUCCUCAACAUAUGCCAAGUACAUGGCAUGCCAAAGAUUAUGGGCGUCCUCACGCAUUUGGAUAUGAUCAAGAAUCCCAAAC ..............((((((....(((......(((((...)))))..(((((((......))))))))))..(((((((.((((....)))).......)))))))..))))))..... ( -29.60, z-score = -0.39, R) >droVir3.scaffold_13049 15084661 120 - 25233164 GUGAUGCCAGCUAUGGCUUUGAAAUGGAGAUCUUCGAGUUCCUCAAUAUAUGCCAGGUACACGGCAUGCCCAAGAUAAUGGGUGUGCUGACGCAUCUGGACAUGAUCAAGAAUCCCAAAC .....((((....)))).......(((.((((((.(((...)))....((((((((((.(.(((((((((((......)))))))))))..).)))))).))))...))).))))))... ( -39.90, z-score = -2.32, R) >droMoj3.scaffold_6680 10646982 120 + 24764193 GUGAUGCAAGCUAUGGCUUUGAGAUGGAAAUCUUCGAGUUCCUGAACAUUUGCCAGGUACAUGGCAUGCCCAAGAUCAUGGGCGUGCUAACCCAUUUGGAUAUGAUCAAGAAUCCAAAGC .........(((.((((((.(((((....))))).)))(((.(((.(((...((((((...(((((((((((......)))))))))))....))))))..))).))).))).))).))) ( -42.10, z-score = -2.92, R) >droGri2.scaffold_15110 3851185 120 - 24565398 GCGAUGCCAGCUACGGCUUCGAAAUGGAGAUUUUUGAGUUUCUCAACAUUUGCCAGGUGCACGGCAUGCCCAAGAUUAUGGGCGUACUUACGCACCUGGACAUGAUCAAGAAUCCGAAAC .(((.(((......))).))).....(.((((((((((((....)))...((((((((((..((.(((((((......))))))).))...)))))))).))...))))))))))..... ( -44.50, z-score = -4.19, R) >anoGam1.chr2L 22139950 120 + 48795086 UCGACGCUAGCUUCGGCUUCGAGAUGGAGGUGUUUGAGUUUCUGAACAUUUGCCAGGUGCACGGAAUGCCGAAGAUUAUGGGCAUCCUGAACCAUCUGGACGUGAUCAAGAACGCGAAGG ..........((((((((((......)))))......(((..(((.(((...(((((((..(((.(((((.(......).))))).)))...)))))))..))).)))..))).))))). ( -39.20, z-score = -1.13, R) >apiMel3.Group10 2577205 120 + 11440700 UAGAUGCAUCUUUUGGUUUUGAGAUGGAAAUAUUUGAAUUUUUAAAUAUUUGUCAGGUUCAUGGUAUGCCUAGAAUUAUGGGUGUUUUAACUCAUUUAGAUUUAAUAAAAAAUGUUAAGC ..(((.((((((........)))))).((((((((((....))))))))))))).(((((..((....))..)))))((((((......)))))).....(((((((.....))))))). ( -22.00, z-score = -0.72, R) >triCas2.ChLG8 6613526 120 + 15773733 GUGACGCGAGUUUCGGCUUCGAAAUGGAAGUGUUCGAGUUUUUAAACAUUUGCCAAGUCCAUGGAAUGCCCAAAAUCAUGGGCAUCUUGACCCAUCUAGACAUGAUCAAAAACGCGAAAA ....((((.((((((....))))))(((((((((..(....)..))))))).))..(((.((((.(((((((......)))))))......))))...)))...........)))).... ( -31.30, z-score = -0.99, R) >consensus GCGACGCAAGCUACGGCUUCGAAAUGGAGAUCUUUGAGUUCCUCAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAAUCCCAAGC .(((.((........)).)))................((((.....(((...((((((....((.(((((((......))))))).)).....))))))..))).....))))....... (-23.28 = -21.67 + -1.61)
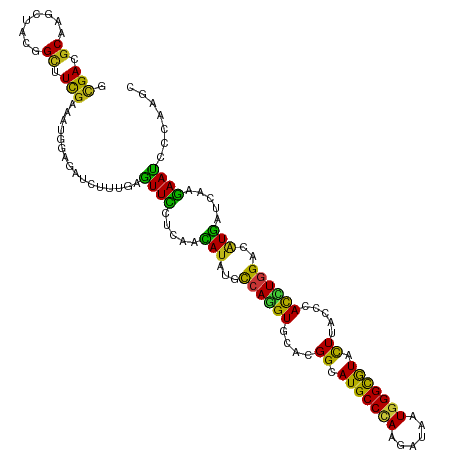
| Location | 17,010,253 – 17,010,373 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.40 |
| Shannon entropy | 0.36963 |
| G+C content | 0.50298 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -19.88 |
| Energy contribution | -19.65 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 17010253 120 + 24543557 CCUGAACAUCUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGCAGCUGAGGAAACGAAAGAAGGAACUGAAGCACCGCUUUUG ..(((.(((...(((((((...((.(((((((......))))))).))....)))))))..))).)))........((((.(((..((...(....)......))..)))...))))... ( -33.80, z-score = -1.53, R) >droSim1.chr3L 16352347 120 + 22553184 CCUGAACAUCUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGCAGCUGAGAAAGCGAAAGAAGGAGCUCAAGCACCGCUUCUG ..(((.(((...(((((((...((.(((((((......))))))).))....)))))))..))).)))........((((.(((.....))).......((.((....)).))))))... ( -38.80, z-score = -2.33, R) >droSec1.super_0 9070848 120 + 21120651 CCUGAACAUCUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGCAGCUGAGGAAGCGAAAGAAGGAGCUCAAGCACCGCUUCUG ..(((.(((...(((((((...((.(((((((......))))))).))....)))))))..))).)))........((((.(((.....))).......((.((....)).))))))... ( -38.80, z-score = -1.75, R) >droYak2.chr3L 7133736 120 - 24197627 CCUAAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCAUCUGGACAUGAUCAAGAACCCCAAACAGCUGAGGAAGCGGAAGAAGGAGCUAAAGCAUCGUUUUUG ((......(((((((((((...((.(((((((......))))))).))....))))))).)))).................(((.....)))))(((((.((((....)).)).))))). ( -34.30, z-score = -1.53, R) >droEre2.scaffold_4784 8929520 120 - 25762168 CCUGAACAUCUGCCAAGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAACAGCUGAGAAAGCGAAAGAAGGAGCUGAAGCACCGCUUUUG ..(((.(((...(((.(((...((.(((((((......))))))).))....))).)))..))).))).............(((.....)))(((((..((.((....)).)).))))). ( -30.70, z-score = -1.19, R) >droAna3.scaffold_13337 9684657 120 - 23293914 CCUCAACAUAUGCCAGGUGCAUGGAAUGCCCAAAAUCAUGGGUGUCCUCACUCACCUGGACAUGAUUAAGAAUCCCAAGCAGCUGCGCAAAAGGAAGAAGGAACUGAAGCAUCGUUUCUG (((.....(((((((((((...(((.((((((......))))))))).....))))))).))))..............((....)).....)))....((((((.((....)))))))). ( -33.50, z-score = -0.85, R) >dp4.chrXR_group8 8513049 120 - 9212921 CCUCAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUCAUGGGCGUGCUCACCCACCUGGACAUGAUCAAGAAUCCCAAGCAGCUGCGCAAGCGCAAGAAGGAGCUGAAGCACCGCUUCUG ........(((((((((((...((((((((((......))))))))))....))))))).))))............((((.((.((....)))).....((.((....)).))))))... ( -48.40, z-score = -3.14, R) >droPer1.super_32 784429 120 + 992631 CCUCAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUCAUGGGCGUGCUCACCCACCUGGACAUGAUCAAGAAUCCCAAGCAGCUGCGCAAGCGCAAGAAGGAGCUGAAGCACCGCUUCUG ........(((((((((((...((((((((((......))))))))))....))))))).))))............((((.((.((....)))).....((.((....)).))))))... ( -48.40, z-score = -3.14, R) >droWil1.scaffold_180955 216256 120 + 2875958 CCUCAACAUAUGCCAAGUACAUGGCAUGCCAAAGAUUAUGGGCGUCCUCACGCAUUUGGAUAUGAUCAAGAAUCCCAAACAAUUGAGGAAGCGCAAAAAGGAACUGAAACAUCGAUUCUG ........(((((((......)))))))..........((.((.((((((....(((((...............)))))....)))))).)).))....((((.(((....))).)))). ( -27.96, z-score = -0.90, R) >droVir3.scaffold_13049 15084701 120 - 25233164 CCUCAAUAUAUGCCAGGUACACGGCAUGCCCAAGAUAAUGGGUGUGCUGACGCAUCUGGACAUGAUCAAGAAUCCCAAACAGCUGCGCAAACGCAAAAAGGAACUGAAGCAUCGCUUCUG (((.....((((((((((.(.(((((((((((......)))))))))))..).)))))).))))...................((((....))))...)))....(((((...))))).. ( -37.60, z-score = -2.39, R) >droMoj3.scaffold_6680 10647022 120 + 24764193 CCUGAACAUUUGCCAGGUACAUGGCAUGCCCAAGAUCAUGGGCGUGCUAACCCAUUUGGAUAUGAUCAAGAAUCCAAAGCAACUUCGCAAACGGAAGAAGGAGCUGAAGCAUCGAUUCUG ..(((.(((...((((((...(((((((((((......)))))))))))....))))))..))).)))((((((...(((..((((..........))))..)))((....)))))))). ( -40.10, z-score = -2.99, R) >droGri2.scaffold_15110 3851225 120 - 24565398 UCUCAACAUUUGCCAGGUGCACGGCAUGCCCAAGAUUAUGGGCGUACUUACGCACCUGGACAUGAUCAAGAAUCCGAAACAAUUGCGCAAGCGCAAAAAAGAGCUCAAGCAUCGCUUCUG ......(((...((((((((..((.(((((((......))))))).))...))))))))..)))....((((..(((.....(((((....)))))......((....)).))).)))). ( -40.80, z-score = -3.36, R) >anoGam1.chr2L 22139990 120 + 48795086 UCUGAACAUUUGCCAGGUGCACGGAAUGCCGAAGAUUAUGGGCAUCCUGAACCAUCUGGACGUGAUCAAGAACGCGAAGGCGCUGAAAAUGCAGAAAAAGGUACUGAAGCAUCGCUUCUG (((((.(((...(((((((..(((.(((((.(......).))))).)))...)))))))..))).)).))).(((....)))(((......)))...........(((((...))))).. ( -35.30, z-score = -1.34, R) >triCas2.ChLG8 6613566 120 + 15773733 UUUAAACAUUUGCCAAGUCCAUGGAAUGCCCAAAAUCAUGGGCAUCUUGACCCAUCUAGACAUGAUCAAAAACGCGAAAACUCUCAAAAACACGAAAAAAGUCCUAAAACACCGGUUCUG ........(((((...(((...(((.((((((......))))))))).)))..(((.......))).......))))).....................((..((........))..)). ( -17.50, z-score = 0.05, R) >consensus CCUCAACAUAUGCCAGGUGCACGGCAUGCCCAAGAUAAUGGGCGUACUUACCCACCUGGACAUGAUCAAGAACCCCAAGCAGCUGAGCAAGCGCAAGAAGGAGCUGAAGCACCGCUUCUG ......(((...((((((....((.(((((((......))))))).)).....))))))..))).........................................(((((...))))).. (-19.88 = -19.65 + -0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:35 2011