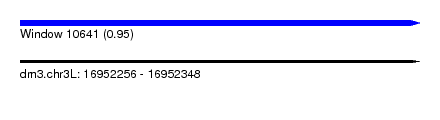
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,952,256 – 16,952,348 |
| Length | 92 |
| Max. P | 0.950611 |

| Location | 16,952,256 – 16,952,348 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Shannon entropy | 0.67917 |
| G+C content | 0.49978 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
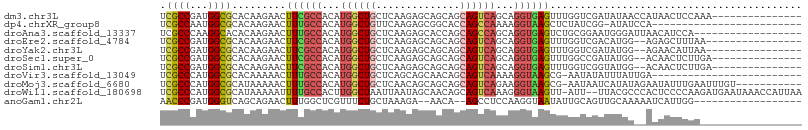
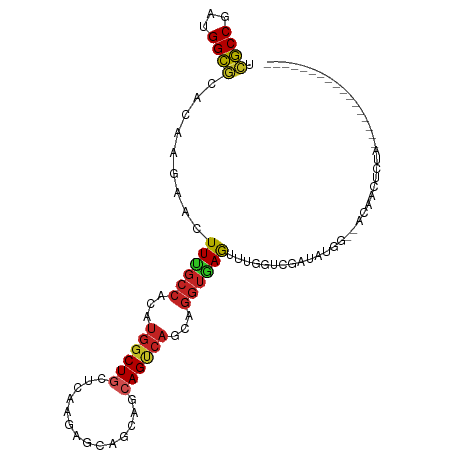
>dm3.chr3L 16952256 92 + 24543557 UCGCCGAUGGCGCACAAGAACUUCGCCACAUGGCUGCUCAAGAGCAGCAGCAGUCAGCAGGUGAGUUUGGUCGAUAUAACCAUAACUCCAAA--------------- ..((((.(((((...........))))))...((((((....)))))).((.....)).)))(((((((((.......)))).)))))....--------------- ( -27.70, z-score = -1.19, R) >dp4.chrXR_group8 4774180 81 + 9212921 UCGCCAAUGGCGCACAAGAACUUUGCCACAUGGCUGUUCAAGAGCGGCACCAGCCAAAAGGUAAGCUCUAUCGG-AUAUCCA------------------------- .((((...)))).....((((...(((....))).)))).(((((...(((........)))..)))))...((-....)).------------------------- ( -24.70, z-score = -1.55, R) >droAna3.scaffold_13337 9623776 89 - 23293914 UCGCCCAAGGCACACAAGAACUUUGCCACAUGGCUGCUCAAGAGCACCAGCAGCCAGCAGGUGAGUCUGCGGAAUGGGAUUAACAUCCA------------------ ...((((.((((...........))))...((((((((..........))))))))(((((....)))))....))))...........------------------ ( -28.40, z-score = -0.85, R) >droEre2.scaffold_4784 8871652 89 - 25762168 UCGCCGAUGGCGCACAAGAACUUCGCCACAUGGCUGCUCAAGAGCAGCAGCAGUCAGCAGGUGAGUUUGGUCGACAUGG--AGAGCUUUAA---------------- ((.((.(((.((.((..(((((.((((.....((((((....)))))).((.....)).))))))))).)))).)))))--.)).......---------------- ( -29.10, z-score = -0.40, R) >droYak2.chr3L 7074456 89 - 24197627 UCGCCGAUGGCGCACAAGAACUUCGCCACAUGGCUGCUCAAGAGCAGCAGCAGUCAGCAGGUGAGUUUGGUCGAUAUGG--AGAACAUUAA---------------- ((.((.(((.((.((..(((((.((((.....((((((....)))))).((.....)).))))))))).)))).)))))--.)).......---------------- ( -27.30, z-score = -0.67, R) >droSec1.super_0 9013664 90 + 21120651 UCGCCGAUGGCGCACAAGAACUUCGCCACAUGGCUGCUCAAGAGCAGCAGCAGUCAGCAGGUGAGUUUGGCCGAUAUGG--ACAACUCUUGA--------------- .((((...))))..(((((..((((((.....((((((....)))))).((.....)).)))))).(((.((.....))--.))).))))).--------------- ( -29.70, z-score = -0.40, R) >droSim1.chr3L 16291013 90 + 22553184 UCGCCGAUGGCGCACAAGAACUUCGCCACAUGGCUGCUCAAGAGCAGCAGCAGUCAGCAGGUGAGUUUGGUCGGUAUGG--ACAACUCUUGA--------------- .((((...))))..(((((..((((((.....((((((....)))))).((.....)).))))))....(((......)--))...))))).--------------- ( -30.60, z-score = -0.58, R) >droVir3.scaffold_13049 15020851 81 - 25233164 UCGCCCAUGGCGCACAAAAACUUUGCCACAUGGCUGCUCAGCAGCAACAGCAGUCAAAAGGUAAGCG-AAUAUAUUUAUUGA------------------------- ((((((.(((((...........)))))..((((((((..........))))))))...))...)))-).............------------------------- ( -20.00, z-score = -0.55, R) >droMoj3.scaffold_6680 10585148 95 + 24764193 UCGCCCAUGGCGCAUAAAAACUUUGCCACAUGGCUGCUCAACAGCAGCAGCAGUCAGAAGGUAAGCG-AAUAAUCAUAUAGAAUAUUUGAAUUUGU----------- .((((...)))).........((((((...(((((((((.......).))))))))...))))))((-((((.((.....)).)))))).......----------- ( -24.60, z-score = -1.40, R) >droWil1.scaffold_180698 1290509 104 + 11422946 UCGCCCAUGGCGCAUAAAAAUUUUGCCACUUGGCUAAUUAAUAGCAACAGCAGUCAAAGGGUAAGUU-AUU--UUACGCCCACUCCCCAAGAUGAAUAAACCAUUAA .......(((.((.(((((..((((((..((((((........((....))))))))..))))))..-.))--))).)))))........((((.......)))).. ( -19.70, z-score = -0.31, R) >anoGam1.chr2L 18835561 85 - 48795086 AACCCGAUGGGUCAGCAGAACUUUGGCUCGUUUCUGCUAAAGA--AACA--AGCCUCCAAGGUAAUAUUGCAGUUGCAAAAAUCAUUGG------------------ ...(((((((.........(((((((((.((((((.....)))--))).--))))...)))))....((((....))))...)))))))------------------ ( -24.80, z-score = -2.25, R) >consensus UCGCCGAUGGCGCACAAGAACUUUGCCACAUGGCUGCUCAAGAGCAGCAGCAGUCAGCAGGUGAGUUUGGUCGAUAUGG__ACAACUCUA_________________ .((((...)))).........((((((...((((((..............))))))...)))))).......................................... (-14.95 = -14.93 + -0.02)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:25 2011