
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,483,090 – 5,483,203 |
| Length | 113 |
| Max. P | 0.706273 |

| Location | 5,483,090 – 5,483,203 |
|---|---|
| Length | 113 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.27 |
| Shannon entropy | 0.18753 |
| G+C content | 0.47428 |
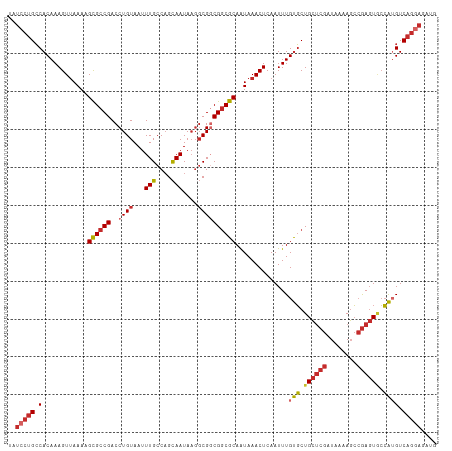
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -25.93 |
| Energy contribution | -26.97 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.706273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5483090 113 + 23011544 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGGAGAUG ..(((((.((...((((....((((((.((.((...(((....)))....))))))))))....)))).....((.((.(((((........))))))))))).))))).... ( -34.60, z-score = -1.61, R) >droSim1.chr2L 5285831 113 + 22036055 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGGAGAUG ..(((((.((...((((....((((((.((.((...(((....)))....))))))))))....)))).....((.((.(((((........))))))))))).))))).... ( -34.60, z-score = -1.61, R) >droSec1.super_5 3556982 113 + 5866729 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGAAGAUG ....(((.((...((((....((((((.((.((...(((....)))....))))))))))....)))).....((.((.(((((........))))))))))).)))...... ( -29.70, z-score = -0.56, R) >droEre2.scaffold_4929 5567206 113 + 26641161 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGGAGAUG ..(((((.((...((((....((((((.((.((...(((....)))....))))))))))....)))).....((.((.(((((........))))))))))).))))).... ( -34.60, z-score = -1.61, R) >droYak2.chr2L 8611182 113 - 22324452 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGGAGAUG ..(((((.((...((((....((((((.((.((...(((....)))....))))))))))....)))).....((.((.(((((........))))))))))).))))).... ( -34.60, z-score = -1.61, R) >droAna3.scaffold_12916 6512525 113 - 16180835 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGGAGAUA ..(((((.((...((((....((((((.((.((...(((....)))....))))))))))....)))).....((.((.(((((........))))))))))).))))).... ( -34.60, z-score = -1.59, R) >dp4.chr4_group1 4792995 109 + 5278887 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGUCGGCGCAGUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUACCAAGUCAGGA---- ..(((((((((((((((.(..((((((((((.....(((....)))...)).))))))))..).))))....))))).((((((........))))))....).)))))---- ( -36.20, z-score = -3.32, R) >droPer1.super_5 6389778 109 - 6813705 UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGUCGGCGCAGUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUACCAAGUCAGGA---- ..(((((((((((((((.(..((((((((((.....(((....)))...)).))))))))..).))))....))))).((((((........))))))....).)))))---- ( -36.20, z-score = -3.32, R) >droWil1.scaffold_180708 648802 102 + 12563649 UAUCCUGGCACAAAGUAAAAUGUGCCGACCUGUAAUUUGCCUGUAAUA---CGGCGUCGCAAUAAACUCAAUUUGCUCUGACCGAUAAAAUAAAACUCGAGUGUC-------- ((((..((((((........))))))((((((((..(((....)))))---))).)))((((..........)))).......))))..................-------- ( -20.30, z-score = -1.29, R) >consensus UAUCCUGCCACAAAGUUAAAAGCGCCGACCUGUAAUUUGCCAGCAAUAAGGCGGCGGCGCAAUAAACUCAAUUUGUGCUGCUCGAUAAAAGCCGAGUGCCAUGUCAGGAGAUG ..(((((.(............((((((..((((...(((....)))....))))))))))..............(((.((((((........)))))).)))).))))).... (-25.93 = -26.97 + 1.04)

| Location | 5,483,090 – 5,483,203 |
|---|---|
| Length | 113 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.27 |
| Shannon entropy | 0.18753 |
| G+C content | 0.47428 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -27.10 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5483090 113 - 23011544 CAUCUCCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((.(((.((.(((((......)))))..)).......(((((....(((((((((((........)))........)).))))))....))))).))).))))).. ( -36.00, z-score = -1.60, R) >droSim1.chr2L 5285831 113 - 22036055 CAUCUCCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((.(((.((.(((((......)))))..)).......(((((....(((((((((((........)))........)).))))))....))))).))).))))).. ( -36.00, z-score = -1.60, R) >droSec1.super_5 3556982 113 - 5866729 CAUCUUCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((.(((.((.(((((......)))))..)).......(((((....(((((((((((........)))........)).))))))....))))).))).))))).. ( -33.10, z-score = -0.88, R) >droEre2.scaffold_4929 5567206 113 - 26641161 CAUCUCCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((.(((.((.(((((......)))))..)).......(((((....(((((((((((........)))........)).))))))....))))).))).))))).. ( -36.00, z-score = -1.60, R) >droYak2.chr2L 8611182 113 + 22324452 CAUCUCCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((.(((.((.(((((......)))))..)).......(((((....(((((((((((........)))........)).))))))....))))).))).))))).. ( -36.00, z-score = -1.60, R) >droAna3.scaffold_12916 6512525 113 + 16180835 UAUCUCCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((.(((.((.(((((......)))))..)).......(((((....(((((((((((........)))........)).))))))....))))).))).))))).. ( -36.00, z-score = -1.63, R) >dp4.chr4_group1 4792995 109 - 5278887 ----UCCUGACUUGGUACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUACUGCGCCGACGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ----(((((........(((((......)))))...((((....(((((....(((((((((((........))).........))))))))....))))))))).))))).. ( -36.70, z-score = -2.49, R) >droPer1.super_5 6389778 109 + 6813705 ----UCCUGACUUGGUACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUACUGCGCCGACGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ----(((((........(((((......)))))...((((....(((((....(((((((((((........))).........))))))))....))))))))).))))).. ( -36.70, z-score = -2.49, R) >droWil1.scaffold_180708 648802 102 - 12563649 --------GACACUCGAGUUUUAUUUUAUCGGUCAGAGCAAAUUGAGUUUAUUGCGACGCCG---UAUUACAGGCAAAUUACAGGUCGGCACAUUUUACUUUGUGCCAGGAUA --------(((((((((((((((((.....))).)))))...))))))..........(((.---.......))).........)))((((((........))))))...... ( -23.80, z-score = -1.47, R) >consensus CAUCUCCUGACAUGGCACUCGGCUUUUAUCGAGCAGCACAAAUUGAGUUUAUUGCGCCGCCGCCUUAUUGCUGGCAAAUUACAGGUCGGCGCUUUUAACUUUGUGGCAGGAUA ....(((((........(((((......)))))...((((....(((((....(((((((((((........)))........)).))))))....))))))))).))))).. (-27.10 = -28.08 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:10 2011