
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,922,992 – 16,923,111 |
| Length | 119 |
| Max. P | 0.568361 |

| Location | 16,922,992 – 16,923,111 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 77.37 |
| Shannon entropy | 0.41138 |
| G+C content | 0.39061 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.568361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16922992 119 + 24543557 ACAUCAGAU-AGGCCUAUGUUCUUAACAUAGGAACAUAGGUCAGUU-GGGAGGAGGAACUA------CCAAUUAGAUUCUGUUGGGUAACCUACUACA----UAGAGCCCAUUUUCAGCAACUGAUUUCAU ..(((((..-.((((((((((((((...)))))))))))))).(((-(..((..((..(((------(((((........)))))(((......))).----)))..))..))..))))..)))))..... ( -41.50, z-score = -4.62, R) >droSim1.chr3L 16270567 119 + 22553184 ACAUUAGAA-AGGCCUUUGUUCUUAACAUAGGAACAUAGGUCAGUU-GGGAGGAGGAACUA------CCAAUUAGAUUCUGUUGGAUAACCUACAAUA----UAGAGCCCAUAUUCAGCAACUGAUUUCUU ..(((((..-.(((((.((((((((...)))))))).)))))((((-((.((......)).------))))))......(((((((((.((((.....----))).)....))))))))).)))))..... ( -30.30, z-score = -1.72, R) >droSec1.super_0 8993362 119 + 21120651 ACAUUAGAA-AGGCCUUUGUUCUUAACAUAGGAACAUAGGUCAGUU-GGGAGGAGGAACUA------CCAAUUAGAUUCUGUUGGAUAACCUACAAUA----UAGAGCCCAUUUUCAGCAACUGAUUUCUU ..(((((..-.(((((.((((((((...)))))))).))))).(((-(..((..((..(((------(((((........))))).............----)))..))..))..))))..)))))..... ( -31.91, z-score = -2.20, R) >droYak2.chr3L 7053813 118 - 24197627 ACUUUAGUUUGGGCCUCUGUUCUUAACAUAGGAACGUAGGUCAGUU-GGGAGGAGGAACUA------CCACUUAGAUUCACUUGGAUAAACUACGA------AAUAGCCCACUUUCAGCCACCGAUUUCUU ........((((((((.((((((((...)))))))).))))..(((-(..((..((.....------........((((....)))).........------.....))..))..))))..))))...... ( -27.73, z-score = -0.03, R) >droEre2.scaffold_4784 8851337 121 - 25762168 ACAUUGGUUUAGGCCUCUGUUCUUAACGUAGGAACAUUGGUCAGUU-GGGAAGAGGAACUA------CC-CUUAGAUUCUGUUGGAUAACCUACAAGUAU--AAUAGCCCACUUUCAGGAACCGAUUUCUU ..((((((((.((((..((((((((...))))))))..))))(((.-(((..((((.....------.)-)))......(((.((....)).))).....--.....)))))).....))))))))..... ( -32.60, z-score = -0.90, R) >droAna3.scaffold_13337 9603540 127 - 23293914 ACAUUU-UAAAGGCCUCUGUUCUUAACAUAGGAACAUGGGGAGUUUUGGGUAAAGGAUCUAACCAUCCCAAUGAGCCUUACAUGGUUACACUAAAUAUAUUUAAGUUUUCUUUUUCAUGGAUUGAUUU--- ......-..(((((.((((((((((...))))))))..((((...((((((.....))))))...))))...))))))).(((((.......((((........))))......))))).........--- ( -26.82, z-score = -0.21, R) >consensus ACAUUAGAA_AGGCCUCUGUUCUUAACAUAGGAACAUAGGUCAGUU_GGGAGGAGGAACUA______CCAAUUAGAUUCUGUUGGAUAACCUACAAUA____AAGAGCCCAUUUUCAGCAACUGAUUUCUU ..(((((....(((((.((((((((...)))))))).))))).....(((....((...........))......((((....))))....................)))...........)))))..... (-17.23 = -17.65 + 0.42)

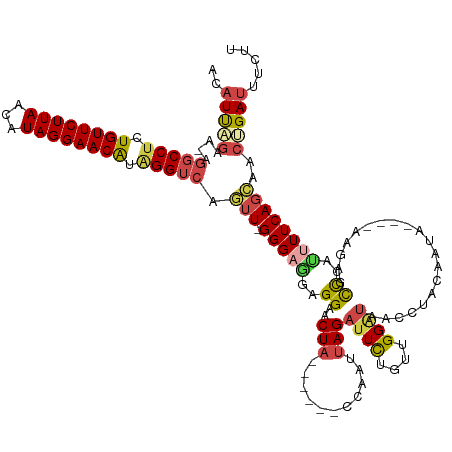
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:23 2011