| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,914,255 – 16,914,354 |
| Length | 99 |
| Max. P | 0.606029 |

| Location | 16,914,255 – 16,914,354 |
|---|---|
| Length | 99 |
| Sequences | 15 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.73 |
| Shannon entropy | 0.67569 |
| G+C content | 0.52898 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.71 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.606029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
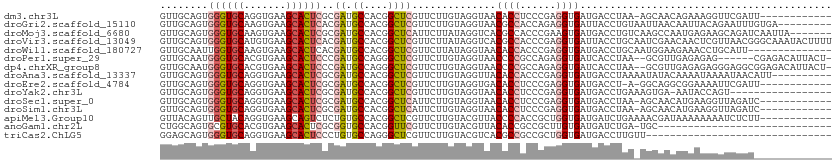
>dm3.chr3L 16914255 99 - 24543557 GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCGUUCUUGUAGGUAACACCUCCCGAGGUGAUGACCUAA-AGCAACAGAAAGGUUCGAUU------------ (((((.((((((((.......))))))))((.((....)))).......(((((..(((((....)))))...))))).-.)))))..............------------ ( -36.10, z-score = -1.98, R) >droGri2.scaffold_15110 13397642 103 - 24565398 GUUGCAGUGGGUGCAAGUGAAGCACUCACGAUGCCACGGCUCGUUCUUGUAGGUAACGCCACCAGAGGUGAUUACCUGUAAUUAACAAUUACAGAAUUUGUGA--------- ((.(((((((((((.......))))))))..))).)).((..(((((...((((((((((......)))).))))))(((((.....))))))))))..))..--------- ( -34.00, z-score = -2.13, R) >droMoj3.scaffold_6680 20771127 105 + 24764193 GUUGCAGUGGGUGCAAGUGAAGCACUCGCGAUGCCACGGCUCAUUCUUAUAGGUCACGCCACCCGAAGUGAUGACCUGUCAAGCCAAUGAGAAGCAGAUCAAUUA------- .((((.((((((((.......))))))))..........((((((((((((((((((((........))).)))))))).)))..))))))..))))........------- ( -35.90, z-score = -2.14, R) >droVir3.scaffold_13049 19578983 112 + 25233164 GUUGCAGUGGGUGCAUGUGAAGCACUCACGAUGCCACGGCUCGUUCUUAUAGGUCACGCCACCCGAGGUGAUUACCUGCAAUCGAACAACUCGUUAACGGGCAAAUACUUUU ((.(((((((((((.......))))))))..))).)).(((((((....(((((..((((......))))...)))))....(((.....)))..))))))).......... ( -35.20, z-score = -1.37, R) >droWil1.scaffold_180727 1883615 98 - 2741493 GUUGCAAUUGGUGCAAGUGAAGCACUCACGAUGCCACGGCUCGUUCUUAUAGGUAACACCACCCGAGGUGAUGACCUGCAAUGGAAGAAACCUGCAUU-------------- ..((((...(((((.......))))).((((.((....))))))(((..(((((..((((......))))...)))))....))).......))))..-------------- ( -26.10, z-score = -0.21, R) >droPer1.super_29 980761 103 - 1099123 GUUGCAAUGGGUGCACGUGAAGCACUCCCGAUGCCAGGGCUCGUUCUUGUAGGUAACCCCGCCAGAGGUGAUCACCUAA--GCGUUGAGAGAG------CGAGACAUUACU- ...(((.(((((((.......)))..)))).)))..(..(((((((((...((....))(((...((((....))))..--)))....)))))------)))).)......- ( -31.30, z-score = -0.21, R) >dp4.chrXR_group8 4738260 109 - 9212921 GUUGCAAUGGGUGCACGUGAAGCACUCCCGAUGCCAGGGCUCGUUCUUGUAGGUAACCCCGCCAGAGGUGAUCACCUAA--GCGUUGAGAGAGGGAGGCGGAGACAUUACU- (((((...((((((.......)))))).(((.((....))))).........))))).(((((..((((....))))..--.(.((....)).)..)))))..........- ( -33.30, z-score = 0.53, R) >droAna3.scaffold_13337 9594343 102 + 23293914 GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCGUUCUUGUAGGUUACACCACCCGAGGUGAUGACCUAAAAUAUACAAAAUAAAAUAACAUU---------- ((((..((((((((.......))))))))((.((....))))((.....(((((((((((......)))).)))))))......)).........))))...---------- ( -32.00, z-score = -2.35, R) >droEre2.scaffold_4784 8842424 98 + 25762168 GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCGUUCUUGUAGGUGACACCUCCCGAGGUGAUGACCU-A-GGCAGGCGGAAAAUUCGAUU------------ .((((.((((((((.......))))))))..(((((((...))).....(((((.((((((....))))).).))))-)-)))).))))...........------------ ( -34.60, z-score = -0.61, R) >droYak2.chr3L 7044273 94 + 24197627 GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCGUUCUUGUAGGUAACACCUCCCGAGGUGAUGACCUGAAAGUGA-AAUACCAGU----------------- ((.(((((((((((.......))))))))..))).))((....(((...(((((..(((((....)))))...))))).....))-)...))...----------------- ( -31.90, z-score = -1.16, R) >droSec1.super_0 8984614 99 - 21120651 GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCAUUCUUGUAGGUAACACCUCCCGAGGUGAUGACCUAA-AGCAACAUGAAGGUUAGAUC------------ (((((.((((((((.......))))))))((.((....)))).......(((((..(((((....)))))...))))).-.)))))..............------------ ( -36.10, z-score = -2.26, R) >droSim1.chr3L 16261896 99 - 22553184 GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCAUUCUUGUAGGUAACACCUCCCGAGGUGAUGACCUAA-AGCAACAUGAAGGUUAGAUC------------ (((((.((((((((.......))))))))((.((....)))).......(((((..(((((....)))))...))))).-.)))))..............------------ ( -36.10, z-score = -2.26, R) >apiMel3.Group10 7985340 100 + 11440700 GUUACAGUUGCUACAGGUGAAGCAGUCUCUGUGCCACGGCUCGUUCUUGUACGUUACCCCACCGCUGGUGAUGAUCUGAAAACGAUAAAAAAAAUCUCUU------------ ...(((((((((.(....).)))))...))))((....))((((((..((.(((((((........)))))))))..))..))))...............------------ ( -19.00, z-score = 0.66, R) >anoGam1.chr2L 14504547 82 - 48795086 CUGGCAGUGCGUGCACGUGAAGCACUCGCGGUGCCACGGUUCGUUCUUGUACGUUACACCGCCGCUUGUGAUGAUCUGA-UGC----------------------------- ..(((.(((.(..(.(((((.....))))))..))))(((.(((......)))....))))))................-...----------------------------- ( -23.70, z-score = 1.46, R) >triCas2.ChLG5 14728002 81 + 18847211 GGAGCAGUGGGUGCAGGUGAAGCACUCCCUGUGCCAGGGCUCGUUCUUGUACGUCACGCCGCCGCUGGUGAUGACCUUGUU------------------------------- .((((....((..((((.((.....))))))..))...))))..........(((((((((....))))).))))......------------------------------- ( -31.00, z-score = -0.33, R) >consensus GUUGCAGUGGGUGCAGGUGAAGCACUCGCGAUGCCACGGCUCGUUCUUGUAGGUAACACCACCCGAGGUGAUGACCUGA_AGCGACAAGAAAAGUAGAU_____________ ........((((((.......))))))..((.((....))))..............((((......)))).......................................... (-13.46 = -13.71 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:22 2011