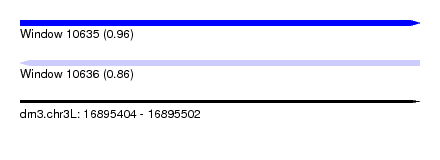
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,895,404 – 16,895,502 |
| Length | 98 |
| Max. P | 0.963062 |

| Location | 16,895,404 – 16,895,502 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 65.00 |
| Shannon entropy | 0.72160 |
| G+C content | 0.52319 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -7.08 |
| Energy contribution | -8.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
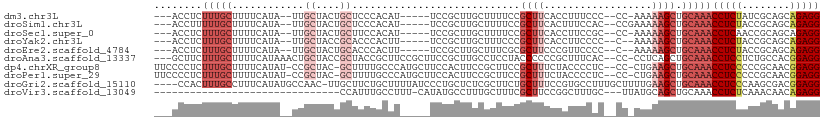
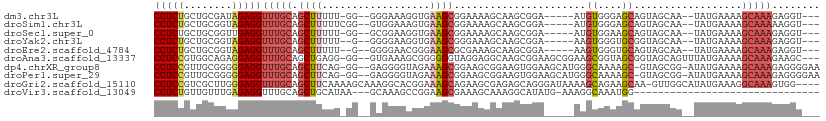
>dm3.chr3L 16895404 98 + 24543557 ---ACCUCUUUGCUUUUCAUA--UUGCUACUGCUCCCACAU-----UCCGCUUGCUUUUCCGCUUCACCUUUCCC--CC-AAAAAGCUGCAAACCUCUAUCGCAGCAGAGG ---.(((((((((........--..((....))........-----...((..((((((................--..-.)))))).))...........)))).))))) ( -17.31, z-score = -2.29, R) >droSim1.chr3L 16243040 99 + 22553184 ---ACCUUUUUGCUUUUCAUA--UUGCUACUGCUCCCACAU-----UCCGCUUGCUUUUCCGCUUCACUUUCCAC--CCGAAAAAGCUGCAAACCUCUACCGCAGCAGAGG ---.(((((((((........--..((....))........-----...((..((((((.((.............--.)).)))))).))...........)))).))))) ( -16.84, z-score = -1.63, R) >droSec1.super_0 8966131 98 + 21120651 ---ACCUCUUUGCUUUUCAUA--UUGCUACUGCUUCCACAU-----UCCGCUUGCUUUUCCGCUUCACCUUCCGC--CC-AAAAAGCUGCAAACCUCAACCGCAGCAGAGG ---.(((((((((........--..((....))........-----...((..((((((..((..........))--..-.)))))).))...........)))).))))) ( -19.60, z-score = -2.41, R) >droYak2.chr3L 7025287 97 - 24197627 ---ACCUCUUUGCUUUUCAUA--UUGCUACCGCACCCACUU-----UCCGCUUGCUUUCCCGCUUCACCUUCCCC--C--AAAAAGCUGCAAACCUCUACCGCAGCAGAGG ---.(((((((((........--.(((....))).......-----...((..(((((.................--.--..))))).))...........)))).))))) ( -16.85, z-score = -2.47, R) >droEre2.scaffold_4784 8824072 97 - 25762168 ---ACCUCUUUGCUUUUCAUA--UUGCUACUGCACCCACUU-----UCCGCUUGCUUUCGCGCUUCCCGUUCCCC--C--AAAAAGCUGCAAACCUCUACCGCAGCAGAGG ---.(((((..((........--.(((....))).......-----...((........))))............--.--.....(((((...........)))))))))) ( -18.30, z-score = -1.69, R) >droAna3.scaffold_13337 9576085 105 - 23293914 ---GCUUCUUUGCUUUUCAUAAACUGCUACCGCUACCGCUUCCGCUUCCGCUUGCCUCCUACCCCCCGCUUUCAC--CC-CCUCAGCUGCAAACCUCCUCUGCCACGGAGG ---.....(((((............((....((....((....))....))..))............(((.....--..-....))).)))))(((((........))))) ( -14.10, z-score = -0.09, R) >dp4.chrXR_group8 4718951 106 + 9212921 UUCCCCUCUUUGCUUUUCAUAU-CCGCUAC-GCUUUUGCCCAUGCUUCCACUUCCGCUUCCGCUUUCUACCCCUC--CC-CUGAAGCUGCAAACCUCCCCCGCAACGGAGG ........(((((.........-..((...-((....))....))..........(((((...............--..-..))))).)))))(((((........))))) ( -17.51, z-score = -2.18, R) >droPer1.super_29 961460 106 + 1099123 UUCCCCUCUUUGCUUUUCAUAU-CCGCUAC-GCUUUUGCCCAUGCUUCCACUUCCGCUUCCGCUUUCUACCCCUC--CC-CUGAAGCUGCAAACCUCCCCCGCAACGGAGG ........(((((.........-..((...-((....))....))..........(((((...............--..-..))))).)))))(((((........))))) ( -17.51, z-score = -2.18, R) >droGri2.scaffold_15110 13378527 106 + 24565398 ----CCACUUUGCCUUUCAUAUGCCAAC-UUGCUUCUGCUUUUAUCCCUGCUCUCGCUUCUGCUUUCCGUGCCUUUGCUUUUGAAGCUGCAAACCUCCCAAGCGACGGAGG ----....(((((...............-..((....))................(((((.((.............))....))))).)))))(((((........))))) ( -18.52, z-score = 0.33, R) >droVir3.scaffold_13049 19556844 76 - 25233164 -------------------------------CCAUUUGCCUUU-CAUAUGCCUUUGCUUUCGCUUCCGGCUUUGC---UUAUGCAGCUGCAAACCUCUCAAACAACAGAGG -------------------------------...(((((....-.....(((...((....))....))).((((---....))))..)))))(((((........))))) ( -16.50, z-score = -1.06, R) >consensus ___ACCUCUUUGCUUUUCAUA__UUGCUACUGCUUCCACUU_____UCCGCUUGCUUUUCCGCUUCCCCUUCCCC__CC_AAAAAGCUGCAAACCUCUACCGCAACAGAGG ........(((((............((....))............................((((..................)))).)))))(((((........))))) ( -7.08 = -8.08 + 1.00)
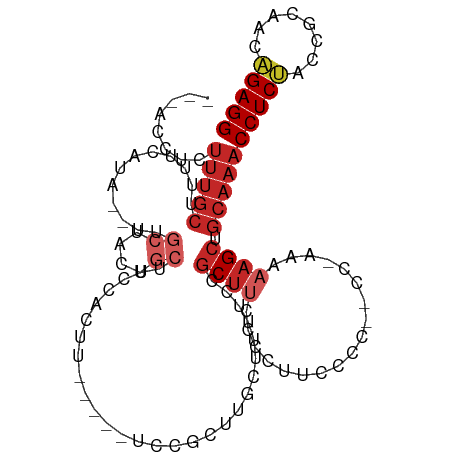
| Location | 16,895,404 – 16,895,502 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.00 |
| Shannon entropy | 0.72160 |
| G+C content | 0.52319 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -9.95 |
| Energy contribution | -10.21 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
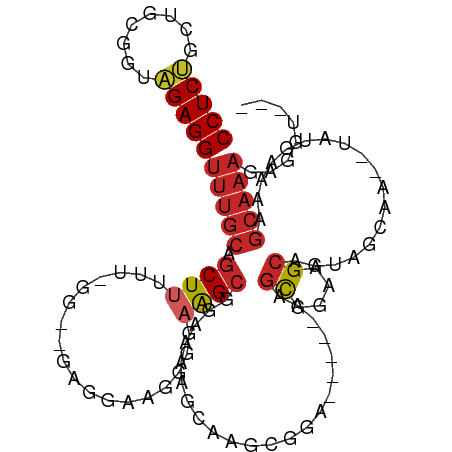
>dm3.chr3L 16895404 98 - 24543557 CCUCUGCUGCGAUAGAGGUUUGCAGCUUUUU-GG--GGGAAAGGUGAAGCGGAAAAGCAAGCGGA-----AUGUGGGAGCAGUAGCAA--UAUGAAAAGCAAAGAGGU--- ((((((((((..((.(..(((((.(((((((-..--..)))))))...((......))..)))))-----.).))...))))).((..--........))..))))).--- ( -25.40, z-score = -1.11, R) >droSim1.chr3L 16243040 99 - 22553184 CCUCUGCUGCGGUAGAGGUUUGCAGCUUUUUCGG--GUGGAAAGUGAAGCGGAAAAGCAAGCGGA-----AUGUGGGAGCAGUAGCAA--UAUGAAAAGCAAAAAGGU--- (((((((....)))))))(((((.(((((((((.--.(........)..)))))))))..((...-----.(((....)))...))..--........))))).....--- ( -29.10, z-score = -1.89, R) >droSec1.super_0 8966131 98 - 21120651 CCUCUGCUGCGGUUGAGGUUUGCAGCUUUUU-GG--GCGGAAGGUGAAGCGGAAAAGCAAGCGGA-----AUGUGGAAGCAGUAGCAA--UAUGAAAAGCAAAGAGGU--- ((((((((((..(((...(((((.(((((((-.(--.(..........)).)))))))..)))))-----...)))..))))).((..--........))..))))).--- ( -26.40, z-score = -0.44, R) >droYak2.chr3L 7025287 97 + 24197627 CCUCUGCUGCGGUAGAGGUUUGCAGCUUUUU--G--GGGGAAGGUGAAGCGGGAAAGCAAGCGGA-----AAGUGGGUGCGGUAGCAA--UAUGAAAAGCAAAGAGGU--- (((((((((((.(.....(((((.(((((((--.--(............).)))))))..)))))-----.....).)))))).((..--........))..))))).--- ( -25.60, z-score = -0.77, R) >droEre2.scaffold_4784 8824072 97 + 25762168 CCUCUGCUGCGGUAGAGGUUUGCAGCUUUUU--G--GGGGAACGGGAAGCGCGAAAGCAAGCGGA-----AAGUGGGUGCAGUAGCAA--UAUGAAAAGCAAAGAGGU--- (((((((((((.(.....(((((.(((((((--(--......))))))))((....))..)))))-----.....).)))))).((..--........))..))))).--- ( -35.20, z-score = -3.62, R) >droAna3.scaffold_13337 9576085 105 + 23293914 CCUCCGUGGCAGAGGAGGUUUGCAGCUGAGG-GG--GUGAAAGCGGGGGGUAGGAGGCAAGCGGAAGCGGAAGCGGUAGCGGUAGCAGUUUAUGAAAAGCAAAGAAGC--- (((((((.(((((.....))))).(((....-.)--))....)))))))(((((..((..((....((....))....))....))..)))))...............--- ( -28.20, z-score = -1.49, R) >dp4.chrXR_group8 4718951 106 - 9212921 CCUCCGUUGCGGGGGAGGUUUGCAGCUUCAG-GG--GAGGGGUAGAAAGCGGAAGCGGAAGUGGAAGCAUGGGCAAAAGC-GUAGCGG-AUAUGAAAAGCAAAGAGGGGAA .((((.((((.......((((((.(((((..-..--............((....)).......))))).((.((....))-.))))))-)).......))))...)))).. ( -28.84, z-score = -2.12, R) >droPer1.super_29 961460 106 - 1099123 CCUCCGUUGCGGGGGAGGUUUGCAGCUUCAG-GG--GAGGGGUAGAAAGCGGAAGCGGAAGUGGAAGCAUGGGCAAAAGC-GUAGCGG-AUAUGAAAAGCAAAGAGGGGAA .((((.((((.......((((((.(((((..-..--............((....)).......))))).((.((....))-.))))))-)).......))))...)))).. ( -28.84, z-score = -2.12, R) >droGri2.scaffold_15110 13378527 106 - 24565398 CCUCCGUCGCUUGGGAGGUUUGCAGCUUCAAAAGCAAAGGCACGGAAAGCAGAAGCGAGAGCAGGGAUAAAAGCAGAAGCAA-GUUGGCAUAUGAAAGGCAAAGUGG---- (((((........)))))(((((.(((((....((....))...).))))....((....))..........)))))..((.-.(((.(........).)))..)).---- ( -25.60, z-score = 0.36, R) >droVir3.scaffold_13049 19556844 76 + 25233164 CCUCUGUUGUUUGAGAGGUUUGCAGCUGCAUAA---GCAAAGCCGGAAGCGAAAGCAAAGGCAUAUG-AAAGGCAAAUGG------------------------------- (((((........)))))(((((.(((.....)---))...(((....((....))...))).....-....)))))...------------------------------- ( -23.50, z-score = -1.66, R) >consensus CCUCUGCUGCGGUAGAGGUUUGCAGCUUUUU_GG__GAGGAAGGGGAAGCGGAAAAGCAAGCGGA_____AAGCGGAAGCAGUAGCAA__UAUGAAAAGCAAAGAGGU___ (((((........)))))(((((.((((..................))))......................((....))..................)))))........ ( -9.95 = -10.21 + 0.26)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:21 2011