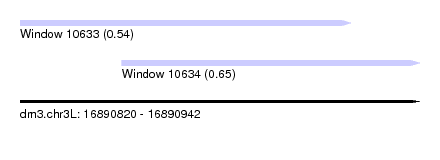
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,890,820 – 16,890,942 |
| Length | 122 |
| Max. P | 0.650438 |

| Location | 16,890,820 – 16,890,921 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.54 |
| Shannon entropy | 0.62338 |
| G+C content | 0.52602 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.09 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.541908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
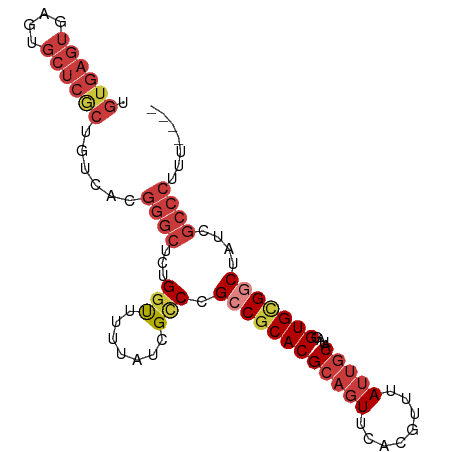
>dm3.chr3L 16890820 101 + 24543557 UGUGUGUCGCCUGGCAAAUCAAGAGUGUCCG-----UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCA-- .(((((.((...(((((((((.((((..(((-----((..(((((....)))))..))))))))))))))).....))))).)))))(((((........))))).-- ( -36.90, z-score = -1.52, R) >droSim1.chr3L 16238494 101 + 22553184 UGUGUGUCGCCUGGCGAAUCGAGAGUGUCCG-----UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCA-- .(((((.((...(((((..(.(((((..(((-----((..(((((....)))))..)))))))))).)......))))))).)))))(((((........))))).-- ( -39.90, z-score = -1.80, R) >droSec1.super_0 8961553 101 + 21120651 UGUGUGUCGCCUGGCGAAUCGAGAGUGUCCG-----UGUGAGUGAAUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCA-- .(((((.((...(((((..(.(((((..(((-----((..(((((....)))))..)))))))))).)......))))))).)))))(((((........))))).-- ( -39.20, z-score = -1.84, R) >droYak2.chr3L 7020715 101 - 24197627 UAUGUGUCGCCUGGCAAACCGAGAGUGUCCG-----UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCCCUUUUAUUGCA-- ...((((.((..((((((((.(((((..(((-----((..(((((....)))))..))))))))))))))).....))).)).))))(((((........))))).-- ( -39.40, z-score = -2.79, R) >droEre2.scaffold_4784 8819553 100 - 25762168 UGUGUGUCGCCUGGCGAAGCGAGAGUGUCCG-----UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCA-GUUUAUUGCA-- (((((((.((..((((((((.(((((..(((-----((..(((((....)))))..)))))))))).)))....))))).)).))))))).....-..........-- ( -42.50, z-score = -2.55, R) >droAna3.scaffold_13337 9571522 105 - 23293914 UGUGUGUCGA-UAGCAGUGCGAGUGUGGUGGAUGGAUGUGAGUGAGUGCUCACUGUCACGGGCUCUGGUUUUUAUCGUCCGCCACACGCAGUUCACAUUUAUUGCA-- .((((((...-..))).)))..(((((((((((((((((((((....)))))).))).(((...)))........))))))))))))(((((........))))).-- ( -37.50, z-score = -0.99, R) >droWil1.scaffold_180727 1854420 103 + 2741493 AACUCAUCAUUCAAUUUAAGUUGUGCGUUUU-----GUUGUGUGUUUUUGUCAUUUAGUUAGUUUUGGUUUUUAUCGCCUGCCGCACGCAGUUCCCUUUAAUGGACUU ...................((.(((((....-----.(((.(((.......))).)))...((...(((.......))).)))))))))(((((........))))). ( -11.40, z-score = 1.77, R) >droVir3.scaffold_13049 19551727 80 - 25233164 -----------------UAAUUG-GCGCUCG-----GCUAUGUCAUCUAGC---UUACGCGGUCUACGCUUUUAUGGCCUGUCGCACGCAUUCACCUUUAAUGACA-- -----------------......-(((..((-----((...(((((..(((---.............)))...)))))..))))..))).................-- ( -15.62, z-score = 0.84, R) >consensus UGUGUGUCGCCUGGCAAAUCGAGAGUGUCCG_____UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCA__ ........................(((..........((((((....)))))).....(((((...(((....)))))))).)))..(((((........)))))... (-13.69 = -14.09 + 0.39)



| Location | 16,890,851 – 16,890,942 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.15 |
| Shannon entropy | 0.33978 |
| G+C content | 0.54198 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -22.47 |
| Energy contribution | -24.47 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16890851 91 + 24543557 UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCAUAGGUGCGGCAAUCGCCCUUU---- .((((((....))))))......((((...(((.......))).((((((((((((........)))))....)))))))....))))...---- ( -34.30, z-score = -1.72, R) >droSim1.chr3L 16238525 91 + 22553184 UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCAUAGGUGCGGCUAUCGCCCUUU---- .((((((....))))))......((((...(((.......))).((((((((((((........)))))....)))))))....))))...---- ( -33.80, z-score = -1.61, R) >droSec1.super_0 8961584 91 + 21120651 UGUGAGUGAAUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCAAAGGUGCGACUAUCGCCCUUU---- .((((((....))))))(((..(((((...(((....))))))))..))).(((((........)))))(((((.(((.....))))))))---- ( -30.80, z-score = -1.41, R) >droYak2.chr3L 7020746 91 - 24197627 UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCCCUUUUAUUGCAUAGGUGCGGCUAUCGCCCUUU---- .((((((....))))))......((((...(((.......))).((((((((((((........)))))....)))))))....))))...---- ( -33.80, z-score = -2.07, R) >droEre2.scaffold_4784 8819584 90 - 25762168 UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCA-GUUUAUUGCAUAGGUGCGGCUAUCGCCCUUU---- .((((((....))))))......((((...(((.......))).((((((((((((...-....)))))....)))))))....))))...---- ( -33.50, z-score = -1.64, R) >droAna3.scaffold_13337 9571557 95 - 23293914 UGUGAGUGAGUGCUCACUGUCACGGGCUCUGGUUUUUAUCGUCCGCCACACGCAGUUCACAUUUAUUGCAUAGGUGCGGCUAUCGCCCUUUUAUU .((((((....))))))......((((...(((....)))(((((((....(((((........)))))...)))).)))....))))....... ( -28.00, z-score = -1.27, R) >droVir3.scaffold_13049 19551744 84 - 25233164 ------UGUCAUCUAGCUUACGCGGUCUACG-CUUUUAUGGCCUGUCGCACGCAUUCACCUUUAAUGACAGAGGUGUGUCCGGCCCAAUCC---- ------...............(((.....))-)......((((.(.(((((.(..(((.......)))..)..))))).).))))......---- ( -19.00, z-score = 0.34, R) >consensus UGUGAGUGAGUGCUCGCUGUCACGGGCUCUGGUUUUUAUCGCCCGCCGCACGCAGUUCACGUUUAUUGCAUAGGUGCGGCUAUCGCCCUUU____ .((((((....))))))......((((...(((.......))).((((((((((((........)))))....)))))))....))))....... (-22.47 = -24.47 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:19 2011