| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,868,744 – 16,868,830 |
| Length | 86 |
| Max. P | 0.993555 |

| Location | 16,868,744 – 16,868,830 |
|---|---|
| Length | 86 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 54.71 |
| Shannon entropy | 0.97022 |
| G+C content | 0.46832 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -3.22 |
| Energy contribution | -3.61 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.11 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
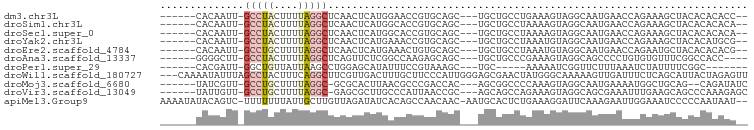
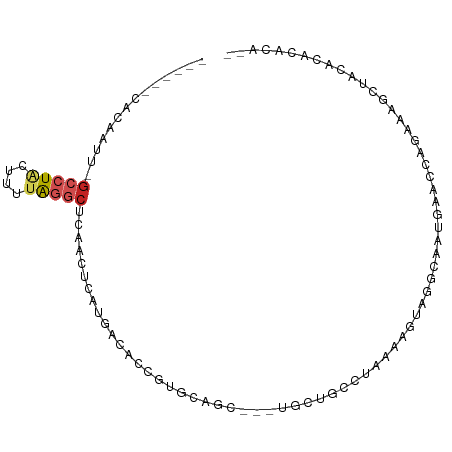
>dm3.chr3L 16868744 86 - 24543557 ------CACAAUU-GCCUACUUUUAGGCUCAACUCAUGGAACCGUGCAGC---UGCUGCCUGAAAGUAGGCAAUGAACCAGAAAGCUACACACACC-- ------....(((-((((((((((((((.((.((((((....)))).)).---))..)))))))))))))))))......................-- ( -34.30, z-score = -5.51, R) >droSim1.chr3L 16216451 86 - 22553184 ------CACAAUU-GCCUACUUUUAGGCUCAACUCAUGGCACCGUGCAGC---UGCUGCCUAAAAGUAGGCAAUGAACCAGAAAGCUACACACACA-- ------....(((-((((((((((((((.((.((((((....)))).)).---))..)))))))))))))))))......................-- ( -34.20, z-score = -5.32, R) >droSec1.super_0 8939218 86 - 21120651 ------CACAAUU-GCCUACUUUUAGGCUCAACUCAUGGCACCGUGCAGC---UGCUGCCUAAAAGUAGGCAAUGAACCAGAAAGCUACACACACA-- ------....(((-((((((((((((((.((.((((((....)))).)).---))..)))))))))))))))))......................-- ( -34.20, z-score = -5.32, R) >droYak2.chr3L 6998404 86 + 24197627 ------CACAAUU-GCCUACUUUUAGGCUCAACUCAUGAAACCGUGCAGC---UGCUGCCUAAAUGUAGGCAAUGAACCAGAAAGCUACACAUGCG-- ------....(((-((((((.(((((((.((.((((((....)))).)).---))..))))))).)))))))))..........((.......)).-- ( -30.10, z-score = -4.42, R) >droEre2.scaffold_4784 8796536 86 + 25762168 ------CACAAUU-GCCUGCUUUUAGGCUCAACUCAUGAAACUGUGCAGC---UGCUGCCUAAAUGUAGGCAAUGAACCAGAAUGCUACACACACG-- ------....(((-((((((.(((((((.((.(((((......))).)).---))..))))))).)))))))))......................-- ( -26.70, z-score = -2.93, R) >droAna3.scaffold_13337 9550354 84 + 23293914 ------GGGGCUU-GCCUACUUUUAGGCUCAGUUCUCGGCCAAGAGCAGC---UGCUGCCCGAAAGUAGGCAGCCCCUGUGUGUUUCGGCCACC---- ------((((((.-(((((((((..(((.((((((((......))).)))---))..)))..))))))))))))))).(((.((....))))).---- ( -46.50, z-score = -4.97, R) >droPer1.super_29 932564 76 - 1099123 ------CACGAUU-GGCUGUUAUUAAGCCUGGAGCAUAUUUCCGUAAAGC---UGC-----AAAAAUCGGUUCUUUAAAUCUAUUUUCGGC------- ------..(((((-((((.......))))((.(((.............))---).)-----)..)))))......................------- ( -13.72, z-score = -0.01, R) >droWil1.scaffold_180727 1827763 95 - 2741493 ---CAAAAUAUUUAGCCUACUUUCAGGCUUCGUUGACUUUGCUUCCCAUUGGGAGCGAACUAUGGGCAAAAAGUUGAUUUCUCAGCAUUACUAGAGUU ---...........(((((...((((......)))).(((((((((....)))))))))...))))).....(((((....)))))............ ( -22.30, z-score = -0.84, R) >droMoj3.scaffold_6680 20717682 85 + 24764193 ------UAUCGUU-GCCUGCUUUUAGGC-GCGCACUUAACGCCCGACCAC---AGCGGCCCCAAAGUAGGCAAUGAAAAUGGCUGCAG--CAGAUAUC ------..(((((-(((((((((..(((-.(((.................---.))))))..)))))))))))))).....((....)--)....... ( -30.67, z-score = -2.36, R) >droVir3.scaffold_13049 19523135 87 + 25233164 ------UAUUGUU-GCCUGCUUUUAGGC-GAGCGCUUGCCCAUUAACCGC---AGCAGCCAGAAAGUAGGCAGCGAAAUUUGAAGCAGCCCAAAGAGC ------..(((((-((((((((((.(((-....(((.((.........))---))).))).)))))))))))))))..((((........)))).... ( -31.50, z-score = -2.87, R) >apiMel3.Group9 2534276 94 + 10282195 AAAAUAUACAGUC-UUUUUUUAUUGCUUGUUAGAUAUCACAGCCAACAAC-AAUGCACUCUGAAAGGAUUCAAAGAAUUGGAAAUCCCCCAAUAAU-- .........((((-(((((....((((((((................)))-)).)))....)))))))))......(((((.......)))))...-- ( -13.79, z-score = -0.63, R) >consensus ______CACAAUU_GCCUACUUUUAGGCUCAACUCAUGACACCGUGCAGC___UGCUGCCUAAAAGUAGGCAAUGAACCAGAAAGCUACACACACA__ ..............(((((....)))))...................................................................... ( -3.22 = -3.61 + 0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:15 2011