| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,846,797 – 16,846,893 |
| Length | 96 |
| Max. P | 0.939371 |

| Location | 16,846,797 – 16,846,893 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 61.03 |
| Shannon entropy | 0.72085 |
| G+C content | 0.45583 |
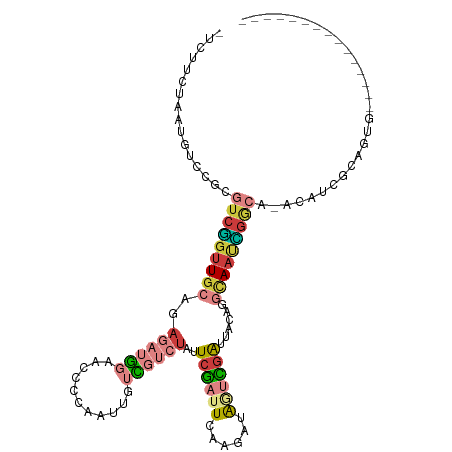
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -10.61 |
| Energy contribution | -11.94 |
| Covariance contribution | 1.33 |
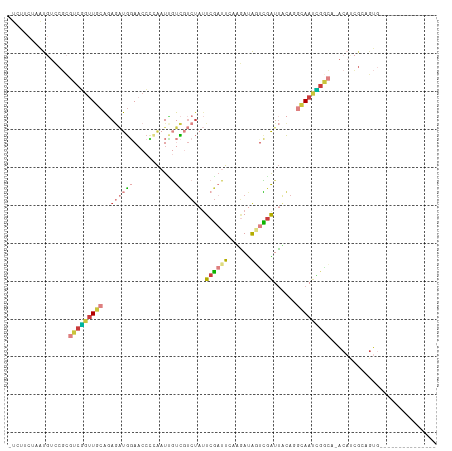
| Combinations/Pair | 1.62 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
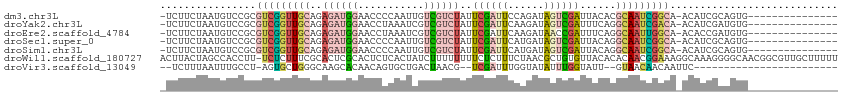
>dm3.chr3L 16846797 96 - 24543557 -UCUUCUAAUGUCCGCGUCGGUUGCAGAGAUGGAACCCCAAUUGUCGUCUAUUCGAUUCCAGAUAGUCGAUUACACGCAAUCGGCA-ACAUCGCAGUG--------------- -.............(((...(((((.((..(((....))).(((.(((.((.((((((......)))))).)).)))))))).)))-))..)))....--------------- ( -25.60, z-score = -1.17, R) >droYak2.chr3L 6976162 96 + 24197627 -UCUUCUAAUGUCCGCGUCGGUUGCAGAGAUGGAACCUAAAUCGUCGUCUAUUCGAUUCAAGAUAGUCGAUUUCAGGCAAUCGACA-ACAUCGAUGUG--------------- -...............(((((((((.(((((.((.........((((......)))).........)).)))))..))))))))).-...........--------------- ( -24.37, z-score = -0.58, R) >droEre2.scaffold_4784 8774184 96 + 25762168 -UCUUCUAAUGUCCGCGUCGGUUGCAGAGAUGGAACCUAAAUCGUCGUCUAUUCGAUUCAAGAUAACCGAUUUCAGGCAAUUGGCA-ACACCGAUGUG--------------- -............((((((((((((...(((....((((((((((..(((..........)))..).)))))).)))..))).)))-..)))))))))--------------- ( -25.20, z-score = -1.09, R) >droSec1.super_0 8917183 96 - 21120651 -UCUUCUAAUGUCCGCGUCGGUUGCAGAGAUGGAACCCCAAUUGUCGUCUAUUCGAUUCAUGAUAGUCGAUUACAGGCAAUCGGCA-ACAUCGCAGUG--------------- -.............(((...(((((.((..(((....))).(((((...((.((((((......)))))).))..))))))).)))-))..)))....--------------- ( -25.20, z-score = -0.75, R) >droSim1.chr3L 16190679 96 - 22553184 -UCUUCUAAUGUCCGCGUCGGUUGCAGAGAUGGAACCCCAAUUGUCGUCUAUUCGAUUCAUGAUAGUCGAUUACAGGCAAUCGGCA-ACAUCGCAGUG--------------- -.............(((...(((((.((..(((....))).(((((...((.((((((......)))))).))..))))))).)))-))..)))....--------------- ( -25.20, z-score = -0.75, R) >droWil1.scaffold_180727 1797519 112 - 2741493 ACUUACUAGCCACCUU-UCUCUUUCGCACUCGCACUCUCACUAUCUUUUUUUUCUCUUUCUAACGCUGUGUUACACACAACGGAAAGGCAAAGGGGCAACGGCGUUGCUUUUU ........(((.((((-(.((((((((....))..............................((.((((.....)))).)))))))).)))))(....)))).......... ( -26.30, z-score = -3.53, R) >droVir3.scaffold_13049 19501158 82 + 25233164 --UCUUUAAUUUGCCU-AGUGCUGGGCAAGCACAACAGUGCUGACUAACG--UCGAUUUGGUAUAUUUGGUAUU--GUAACAACAAUUC------------------------ --.........(((((-(....))))))(((((....)))))(((....)--))((.(((.((((........)--)))....))).))------------------------ ( -20.20, z-score = -1.39, R) >consensus _UCUUCUAAUGUCCGCGUCGGUUGCAGAGAUGGAACCCCAAUUGUCGUCUAUUCGAUUCAAGAUAGUCGAUUACAGGCAAUCGGCA_ACAUCGCAGUG_______________ ................(((((((((..((((((...........))))))..((((((......))))))......)))))))))............................ (-10.61 = -11.94 + 1.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:10 2011