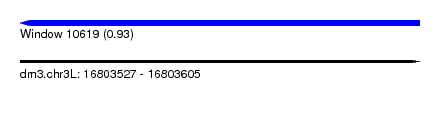
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,803,527 – 16,803,605 |
| Length | 78 |
| Max. P | 0.928740 |

| Location | 16,803,527 – 16,803,605 |
|---|---|
| Length | 78 |
| Sequences | 14 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 75.65 |
| Shannon entropy | 0.49426 |
| G+C content | 0.54937 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -15.07 |
| Energy contribution | -14.91 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
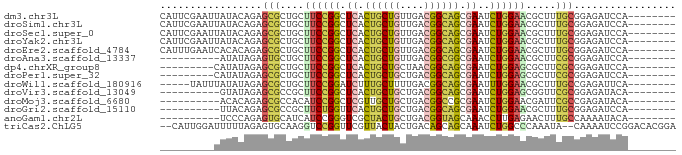
>dm3.chr3L 16803527 78 - 24543557 CAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA-------- .............(((((((....((((((.((.((((((....)))))).))..))))))))))))).((....)).-------- ( -27.80, z-score = -2.06, R) >droSim1.chr3L 16145196 78 - 22553184 CAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA-------- .............(((((((....((((((.((.((((((....)))))).))..))))))))))))).((....)).-------- ( -27.80, z-score = -2.06, R) >droSec1.super_0 8874497 78 - 21120651 CAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA-------- .............(((((((....((((((.((.((((((....)))))).))..))))))))))))).((....)).-------- ( -27.80, z-score = -2.06, R) >droYak2.chr3L 6932629 78 + 24197627 CAUUCGAAUUAUACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA-------- .............(((((((....((((((.((.((((((....)))))).))..))))))))))))).((....)).-------- ( -27.80, z-score = -2.06, R) >droEre2.scaffold_4784 8731525 78 + 25762168 CAUUUGAAUCACACAGAGCGCUGCUUCCGGCUCACUGCUGUUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA-------- ....((.(((.(.(((((((....((((((.((.((((((....)))))).))..))))))))))))).)..))).))-------- ( -28.90, z-score = -2.25, R) >droAna3.scaffold_13337 9488767 68 + 23293914 ----------AUAUAGAGUGCUGCUUCCGGCUCACUGCUGUUGACGGCGGCGAAUCUGGAACGCUUCGCGGAGAUCCA-------- ----------.......(((..((((((((.((.((((((....)))))).))..)))))).))..)))((....)).-------- ( -25.60, z-score = -1.65, R) >dp4.chrXR_group8 732292 69 - 9212921 ---------CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUAACGGCAGCGAAUCUGGAGCGCUUCGCGGAGAUCCA-------- ---------........(((..((((((((.((.((((((....)))))).))..)))))).))..)))((....)).-------- ( -27.10, z-score = -1.30, R) >droPer1.super_32 241758 69 + 992631 ---------CAUAUAGAGCGCUGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAGCGCUUCGCGGAGAUCCA-------- ---------........(((..((((((((.((.((((((....)))))).))..)))))).))..)))((....)).-------- ( -27.10, z-score = -0.95, R) >droWil1.scaffold_180916 336663 73 + 2700594 -----UAUUUAUAUAGAGCGCUGCUUCCGGAUCUUUGCUUUUGACGGCAGCGAAUUUGGAACGCUUUGCCGAGAUUCA-------- -----.((((...(((((((....(((((((((.(((((......))))).).)))))))))))))))...))))...-------- ( -18.50, z-score = -0.62, R) >droVir3.scaffold_13049 8471938 68 - 25233164 ----------GUAUAGAGCGCCGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAGCGGUUCGCGGAGAUACA-------- ----------((((...((((((((((.(..((.((((((....)))))).))..).)))))))..)))....)))).-------- ( -31.90, z-score = -2.66, R) >droMoj3.scaffold_6680 12119920 68 - 24764193 ----------ACACAGAGCGCCACAUCCGGCUCGUUGCUGCUGACGGCCGCGAAUCUGGAACGAUUCGCCGAGAUACA-------- ----------..............(((((((..((.((((....)))).))(((((......))))))))).)))...-------- ( -20.10, z-score = -0.33, R) >droGri2.scaffold_15110 7496914 68 - 24565398 ----------UUACAGAGCGCCGCUUCUGGUUCACUGCUGCUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA-------- ----------...(((((((...((...(((((.((((((....)))))).))))).))..))))))).((....)).-------- ( -26.70, z-score = -2.12, R) >anoGam1.chr2L 26337033 68 - 48795086 ----------UCCCAGAGUGCAUCAUCCGGGUCGCUACUGCUGACGGUAGCAAACCUUGAGAACUUUGCCAAAAUACA-------- ----------...((((((...(((...((((.(((((((....)))))))..)))))))..))))))..........-------- ( -20.30, z-score = -2.18, R) >triCas2.ChLG5 695307 82 + 18847211 --CAUUGGAUUUUUAGAGUGCAAGGUCCGGUUCGUUACUACUGACAGCAGCAAAUCUGGCCCAAAUA--CAAAAUCCGGACACGGA --..(((((((((.....((...((((.((((.(((.((......)).))).)))).))))))....--.)))))))))....... ( -16.70, z-score = 0.25, R) >consensus _________UAUACAGAGCGCUGCUUCCGGCUCACUGCUGCUGACGGCAGCGAAUCUGGAACGCUUUGCGGAGAUCCA________ .................(((....((((((.((.((((((....)))))).))..)))))).....)))................. (-15.07 = -14.91 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:07 2011