| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,479,958 – 5,480,063 |
| Length | 105 |
| Max. P | 0.990777 |

| Location | 5,479,958 – 5,480,063 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.71 |
| Shannon entropy | 0.31688 |
| G+C content | 0.47413 |
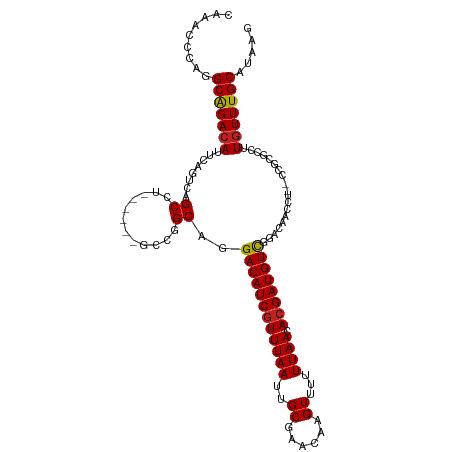
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.09 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
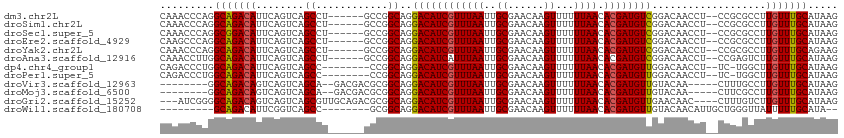
>dm3.chr2L 5479958 105 - 23011544 CAAACCCAGGCAGACAUUCAGUCAGCCU------GCCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU--CCGCGCCUUGUUUGCAUAAG (((((.(((((.(((.....))).))))------)..(((..((((((((((((..((......))...)))).))))))))(((.....)--))..)))..)))))...... ( -35.30, z-score = -4.04, R) >droSim1.chr2L 5282783 105 - 22036055 CAAACCCAGGCAGACAUUCAGUCAGCCU------GCCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU--CCGCGCCUUGUUUGCAUAAG (((((.(((((.(((.....))).))))------)..(((..((((((((((((..((......))...)))).))))))))(((.....)--))..)))..)))))...... ( -35.30, z-score = -4.04, R) >droSec1.super_5 3553877 105 - 5866729 CAAACCCAGGCGGACAUUCAGUCAGCCU------GCCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU--CCGCGCCUUGUUUGCAUAAG (((((.(((((.(((.....))).))))------)..(((..((((((((((((..((......))...)))).))))))))(((.....)--))..)))..)))))...... ( -36.00, z-score = -3.67, R) >droEre2.scaffold_4929 5564045 105 - 26641161 CAAGCCCAGGCAGACAUUCAGUCAGCCU------GCCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU--CCGCGCCUUGUUUGCAUAAG (((((.(((((.(((.....))).))))------)..(((..((((((((((((..((......))...)))).))))))))(((.....)--))..)))..)))))...... ( -35.30, z-score = -3.64, R) >droYak2.chr2L 8608092 105 + 22324452 CAAACCCAGGCAGACAUUCAGUCAGCCU------GCCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU--CCGCGCCUUGUUUGCAGAAG (((((.(((((.(((.....))).))))------)..(((..((((((((((((..((......))...)))).))))))))(((.....)--))..)))..)))))...... ( -35.30, z-score = -3.66, R) >droAna3.scaffold_12916 6509680 105 + 16180835 CAAACCUUGGCAGACAUUCAGUCAGCCU------GCCGGCAGGACAUCAUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU--CCGAGUCUUGUUUGCAUAAG ........(((.(((.....))).)))(------((..((((((((((..((((..((......))...))))...))).(((((.....)--)))))))))))..))).... ( -28.60, z-score = -2.22, R) >dp4.chr4_group1 4789535 102 - 5278887 CAGACCCUGGCAGACAUUCAGUCAGCC--------CCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUUGGACAACCU--UC-UGGCUUGUUUGCAUAAG .........(((((((....(((.((.--------...))..((((((((((((..((......))...)))).)))))))).)))..((.--..-.))..)))))))..... ( -27.40, z-score = -1.48, R) >droPer1.super_5 6386309 102 + 6813705 CAGACCCUGGCAGACAUUCAGUCAGCC--------CCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUUGGACAACCU--UC-UGGCUUGUUUGCAUAAG .........(((((((....(((.((.--------...))..((((((((((((..((......))...)))).)))))))).)))..((.--..-.))..)))))))..... ( -27.40, z-score = -1.48, R) >droVir3.scaffold_12963 19774917 98 - 20206255 --------GGCAGACAGUCAGUCAGCA--GACGACGCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUUGUACAA-----CUUUGCCUUGUUUGCAUAAG --------.((((((((((.(((....--))))))..(((((((((((((((((..((......))...)))).))))))))......-----..))))).)))))))..... ( -31.00, z-score = -2.95, R) >droMoj3.scaffold_6500 8927120 98 + 32352404 --------GGCAGACAGUCAGUCAGCA--GACGACGCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUUGUACAA-----CUUCGCCUUGUUUGCAUAAG --------.((((((((((.(((....--))))))..(((..((((((((((((..((......))...)))).))))))))(.....-----...)))).)))))))..... ( -29.60, z-score = -2.48, R) >droGri2.scaffold_15252 11239600 106 - 17193109 ---AUCGGGGCAGACAGUCAGUCAGCGUUGCAGACGCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUUGAACAAC----CUUUGUCUUGUUUGCAUAAG ---......((((((.....(((.(((((...))))))))((((((((((......))))....(((.((((((.....)))))).)))----...))))))))))))..... ( -29.80, z-score = -1.38, R) >droWil1.scaffold_180708 645069 94 - 12563649 ---------GCAGACAUUCGGUCAGCC--------GCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUUGUACAACAUUGCUGGGUUAUUUUUGCAUA-- ---------((((((.....)))((((--------.((((((((((((((((((..((......))...)))).))))))))(.....).)))))))))).....)))...-- ( -26.90, z-score = -2.35, R) >consensus CAAACCCAGGCAGACAUUCAGUCAGCCU______GCCGGCAGGACAUCGUUUAAUUGCGAACAAGUUUUUUAACACGAUGUCGGACAACCU__CCGCGCCUUGUUUGCAUAAG .........(((((((........((............))..((((((((((((..((......))...)))).))))))))...................)))))))..... (-22.25 = -22.09 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:07 2011