
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,700,784 – 16,700,886 |
| Length | 102 |
| Max. P | 0.749993 |

| Location | 16,700,784 – 16,700,886 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.49405 |
| G+C content | 0.40125 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749993 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
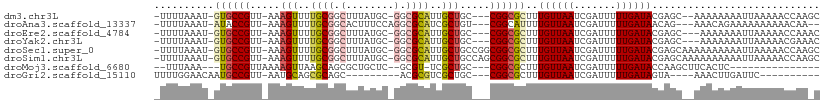
>dm3.chr3L 16700784 102 - 24543557 -UUUUAAAU-GUGCCGUU-AAAGUUUUGCGGCUUUAUGC-GGCGCAUUGCUGC---CGGCGCUUUGUUAAUCGAUUUUUGAUACGAGC--AAAAAAAAUUAAAAACCAAGC -.....(((-((((((((-(((((......)))))).))-))))))))((...---..))((((((((((.......)))))).))))--..................... ( -27.50, z-score = -0.89, R) >droAna3.scaffold_13337 9390443 100 + 23293914 -UUUUAAAU-AUACCGUU-AAAGUUUUGCGGCACUUUCCAGGCGCAUCGCUGU---CGGCAUUUUGUUAAUCGAUUUUUGAUAACAG---AAACAGAAAAAAAAAACAA-- -........-...(((..-..(((..((((.(........).))))..)))..---)))..((((((((.((((...))))))))))---)).................-- ( -19.60, z-score = -0.91, R) >droEre2.scaffold_4784 8628646 101 + 25762168 -UUUUAAAU-GUGCCGUU-AAAGUUUUGCGGCUUUAUGC-GGCGCAUUGCUGC---CGGCGCUUUGUUAAUCGAUUUUUGAUACGAGC---AAAAAAAUUAAAAACCAAAC -.....(((-((((((((-(((((......)))))).))-))))))))((...---..))((((((((((.......)))))).))))---.................... ( -27.50, z-score = -1.37, R) >droYak2.chr3L 6823287 101 + 24197627 -UUUUAAAU-GUGCCGUU-AAAGUUUUGCGGCUUUAUGC-GGCGCAUUGCUGC---CGGCGCUUUGUUAAUCGAUUUUUGAUACGAGC---AAAAAAAUUAAAAACGAAAC -.....(((-((((((((-(((((......)))))).))-))))))))((...---..))((((((((((.......)))))).))))---.................... ( -27.50, z-score = -1.23, R) >droSec1.super_0 8773101 107 - 21120651 -UUUUAAAU-GUGCCGUU-AAAGUUUUGCGGCUUUAUGC-GGCGCAUUGCUGCCGGCGGCGCUUUGUUAAUCGAUUUUUGAUACGAGCAAAAAAAAAAUUAAAAACCAAGC -.....(((-((((((((-(((((......)))))).))-))))))))((((....))))((((((((((.......)))))).))))....................... ( -31.60, z-score = -1.20, R) >droSim1.chr3L 16027609 107 - 22553184 -UUUUAAAU-GUGCCGUU-AAAGUUUUGCGGCUUUAUGC-GGCGCAUUGCUGCCAGCGGCGCUUUGUUAAUCGAUUUUUGAUACGAGCAAAAAAAAAAUUAAAAACCAAGC -.....(((-((((((((-(((((......)))))).))-))))))))((((....))))((((((((((.......)))))).))))....................... ( -31.60, z-score = -1.66, R) >droMoj3.scaffold_6680 12025011 85 - 24764193 --UUUAAA---UGCCGUUAAAAGUUAAGCAGCGCUGCUC--GCGU-UCGCUGC---CGGCGCUUUGUUAAUCGAUUUUUGAUACCAAGCUUCACUC--------------- --......---.((((...........((((((..((..--..))-.))))))---))))((((.((..(((((...))))))).)))).......--------------- ( -20.60, z-score = -0.18, R) >droGri2.scaffold_15110 7400777 84 - 24565398 UUUUGGAACAAUGCCGUU-AAUGCAGCGCAGC---------ACGCGUCGCUGC---CGGCGCUUUGUUAAUCGAUUUUUGAUAGUA----AAACUUGAUUC---------- ......((((((((((..-...((((((..(.---------...)..))))))---)))))..)))))((((((.(((((....))----))).)))))).---------- ( -22.00, z-score = -0.20, R) >consensus _UUUUAAAU_GUGCCGUU_AAAGUUUUGCGGCUUUAUGC_GGCGCAUUGCUGC___CGGCGCUUUGUUAAUCGAUUUUUGAUACGAGC__AAAAAAAAUUAAAAACCAA_C ..........((((((.....(((..((((.(........).))))..))).....))))))..((((((.......))))))............................ (-17.33 = -17.79 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:32:00 2011