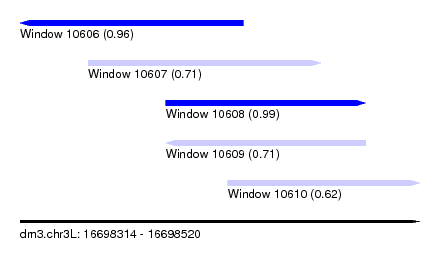
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,698,314 – 16,698,520 |
| Length | 206 |
| Max. P | 0.986603 |

| Location | 16,698,314 – 16,698,429 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.32 |
| Shannon entropy | 0.54222 |
| G+C content | 0.55064 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -13.79 |
| Energy contribution | -15.60 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16698314 115 - 24543557 CUCCUUCGGAAUUAUUUCCUGCCGAAUAACCACUUCCGGAGAAAGCUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCUGCUGCUCCAUCAAUGGAAUUUCG--- ....(((((............))))).......((((((((.......))))))))..........(((((((.(((.(.(((((((........))))))).).))))))))))--- ( -32.00, z-score = -1.81, R) >droAna3.scaffold_13266 10884202 106 + 19884421 ------------CUGCUCCGGCACCGGUACCGCUUCCUCCAGCUCCUGUUCCAGCACGGCCUCCUACACCAACGCCUCCACUGCCGGCACUAUCAAUUCCUCCAUCGUGCCGCCGCCG ------------......((((..((((((.(((......)))..........((.((((......................))))))..................))))))..)))) ( -24.85, z-score = -1.46, R) >droEre2.scaffold_4784 8626245 115 + 25762168 CUCCUUCGGAAUUAUUUCCUCUUGAAAUACCGCCUCCGCAGAAACCUGCUCUGGAUUUACCCCCGUCGGAAUUGCCGCCUGUGGAGCCUUCGGCUGCUCCAUUAAUGCGAUUUCG--- .(((...((...((((((.....))))))...))...((((....))))...)))...........(((((((((.....((((((((....)..)))))))....)))))))))--- ( -34.10, z-score = -3.06, R) >droYak2.chr3L 6820906 115 + 24197627 CUCCUUCGGAAUUAUUUCCUCUGGAAUUUCCACCUCCGCAGAAACCUGCGCUGCAUUUACCCACGUCGGCAUUGCCGCCUGUGGCGCCUCCGGCUGCUCCAUCAGUGCGAUUUCG--- .(((...((((....))))...)))...............((((..(((((((........((.(((((...(((((....)))))...)))))))......))))))).)))).--- ( -31.64, z-score = -1.14, R) >droSec1.super_0 8770674 115 - 21120651 CUCCUUCGGAAAUAUUUCCUGUCGAAUUACCACUUCCGGAGAAAACUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCG--- .......((((....))))..............((((((((.......))))))))..........(((((((((.....(((((((........)))))))....)))))))))--- ( -33.00, z-score = -3.10, R) >droSim1.chr3L 16025160 115 - 22553184 CUCCUUCGGAAAUAUUUCCUGUCGAAUUACCACUUCCGGAGAAAGCUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCUAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCG--- .......((((....))))..............((((((((.......))))))))..........(((((((((.....(((((((........)))))))....)))))))))--- ( -33.00, z-score = -2.44, R) >consensus CUCCUUCGGAAUUAUUUCCUGUCGAAUUACCACUUCCGCAGAAACCUGCUCUGGAAUUACCCCCCUCGGAAUUGCCGCCAGUGGAGCCUCCAGCUGCUCCAUCAAUGCAAUUUCG___ .......((((....))))...............................................(((((((((.....(((((((........)))))))....)))))))))... (-13.79 = -15.60 + 1.81)
| Location | 16,698,349 – 16,698,469 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.92 |
| Shannon entropy | 0.22369 |
| G+C content | 0.49333 |
| Mean single sequence MFE | -39.16 |
| Consensus MFE | -25.08 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16698349 120 + 24543557 UAGCGGCAAUUCCGAGGGGGGUAAUUCCAGAGCAGCUUUCUCCGGAAGUGGUUAUUCGGCAGGAAAUAAUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAG ((((.((..(((((..(..(((............)))..)..))))))).))))(((((.(........).)))))(((..((((((((.(((((....)))))))))).)))..))).. ( -42.80, z-score = -3.35, R) >droEre2.scaffold_4784 8626280 120 - 25762168 AGGCGGCAAUUCCGACGGGGGUAAAUCCAGAGCAGGUUUCUGCGGAGGCGGUAUUUCAAGAGGAAAUAAUUCCGAAGGAGGUAAAUCAACUGGAUGCAAUUCCGGUGGAAGUAAUUCCAA ...(((.....))).(((((....((((((.((((....)))).((.....((((((....((((....))))....))))))..))..))))))....))))).(((((....))))). ( -37.80, z-score = -2.00, R) >droYak2.chr3L 6820941 120 - 24197627 AGGCGGCAAUGCCGACGUGGGUAAAUGCAGCGCAGGUUUCUGCGGAGGUGGAAAUUCCAGAGGAAAUAAUUCCGAAGGAGGUAAACCGACUGGAUGCAAUUCCGGUGGAUGUAAUUCCAG ..((.(((.((((......))))..))).))((((....))))((((((.....((((...((((....))))...))))....))).((((((......)))))).........))).. ( -37.80, z-score = -1.11, R) >droSec1.super_0 8770709 120 + 21120651 UAGCGGCAAUUCCGAGGGGGGUAAUUCCAGAGCAGUUUUCUCCGGAAGUGGUAAUUCGACAGGAAAUAUUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAG ...(((.....)))..((..(((.((((...(.((((..(..(....)..).)))))....)))).)))..))...(((..((((((((.(((((....)))))))))).)))..))).. ( -38.70, z-score = -2.66, R) >droSim1.chr3L 16025195 120 + 22553184 UAGCGGCAAUUCCGAGGGGGGUAAUUCCAGAGCAGCUUUCUCCGGAAGUGGUAAUUCGACAGGAAAUAUUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAG ...(((.....)))..(((..((...(((((...(((((((.((((((((....(((....)))..)))))))).)))))))...))...(((((....))))).)))...))..))).. ( -38.70, z-score = -2.44, R) >consensus UAGCGGCAAUUCCGAGGGGGGUAAUUCCAGAGCAGCUUUCUCCGGAAGUGGUAAUUCGACAGGAAAUAAUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAG ..((.((..(((((.((((((...............))))))))))))).)).........((((....))))...(((..((..((((.(((((....)))))))))...))..))).. (-25.08 = -25.04 + -0.04)

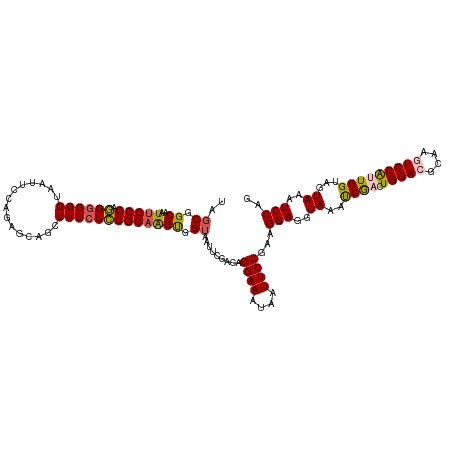
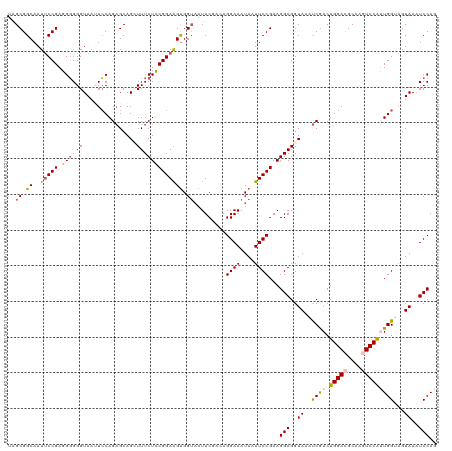
| Location | 16,698,389 – 16,698,492 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.34899 |
| G+C content | 0.45934 |
| Mean single sequence MFE | -35.66 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16698389 103 + 24543557 UCCGGAAGUGGUUAUUCGGCAGGAAAUAAUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUUCCGAGCAAGUU----------------- .(((((........)))))..((((((((((((...(((..((((((((.(((((....)))))))))).)))..)))...)))).)))))))).........----------------- ( -39.10, z-score = -4.86, R) >droEre2.scaffold_4784 8626320 120 - 25762168 UGCGGAGGCGGUAUUUCAAGAGGAAAUAAUUCCGAAGGAGGUAAAUCAACUGGAUGCAAUUCCGGUGGAAGUAAUUCCAAAGGAACUAAUUCCCGGACAGGUUAUACCGGUGGAUGUAAU ((((..(.((((((..(..(.((((.((.((((...((((.((..((.((((((......)))))).))..)).))))...)))).)).)))))......)..)))))).)...)))).. ( -35.10, z-score = -1.85, R) >droYak2.chr3L 6820981 120 - 24197627 UGCGGAGGUGGAAAUUCCAGAGGAAAUAAUUCCGAAGGAGGUAAACCGACUGGAUGCAAUUCCGGUGGAUGUAAUUCCAGAGGAAUUAAUUCCCGAGCAGGUAAAACCGGUGGAGGUAAU (((((((.......)))).(.((((.(((((((...((((.((..((.((((((......))))))))...)).))))...))))))).)))))..)))......(((......)))... ( -37.20, z-score = -2.16, R) >droSec1.super_0 8770749 103 + 21120651 UCCGGAAGUGGUAAUUCGACAGGAAAUAUUUCCGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGGUU----------------- ((.((((..(((..(((....((((....))))...(((..((((((((.(((((....)))))))))).)))..))).)))..)))..)))).)).......----------------- ( -34.30, z-score = -2.96, R) >droSim1.chr3L 16025235 103 + 22553184 UCCGGAAGUGGUAAUUCGACAGGAAAUAUUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACCUAUUCCCGAGCAGGUU----------------- ((.((((..(((..(((....((((....))))...(((..((.(((((.(((((....))))))))))..))..))).)))..)))..)))).)).......----------------- ( -32.60, z-score = -2.37, R) >consensus UCCGGAAGUGGUAAUUCGACAGGAAAUAAUUCCGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGGUU_________________ ...((((..(((..(((....((((....))))...(((..((..((((.(((((....)))))))))...))..))).)))..)))..))))........................... (-21.48 = -21.40 + -0.08)

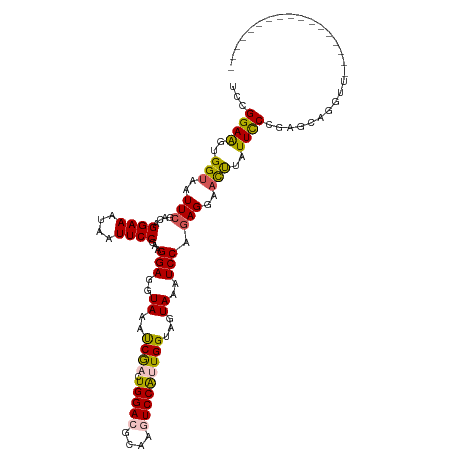
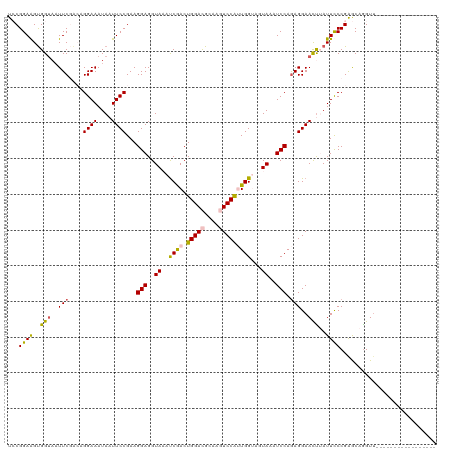
| Location | 16,698,389 – 16,698,492 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.34899 |
| G+C content | 0.45934 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -17.89 |
| Energy contribution | -18.69 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16698389 103 - 24543557 -----------------AACUUGCUCGGAAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUGUACCUCCUUCGGAAUUAUUUCCUGCCGAAUAACCACUUCCGGA -----------------.....((..((((((((.((((...(((..(((......(((((....)))))......)))..)))...)))))))))))).)).......((......)). ( -28.10, z-score = -2.75, R) >droEre2.scaffold_4784 8626320 120 + 25762168 AUUACAUCCACCGGUAUAACCUGUCCGGGAAUUAGUUCCUUUGGAAUUACUUCCACCGGAAUUGCAUCCAGUUGAUUUACCUCCUUCGGAAUUAUUUCCUCUUGAAAUACCGCCUCCGCA ...........((((((.........(((((.(((((((...(((..((..((.((.(((......))).)).))..))..)))...))))))).)))))......))))))........ ( -28.26, z-score = -1.97, R) >droYak2.chr3L 6820981 120 + 24197627 AUUACCUCCACCGGUUUUACCUGCUCGGGAAUUAAUUCCUCUGGAAUUACAUCCACCGGAAUUGCAUCCAGUCGGUUUACCUCCUUCGGAAUUAUUUCCUCUGGAAUUUCCACCUCCGCA ......((((..(((...(((.(((.(((...(((((((..((((......))))..)))))))..)))))).)))..)))......((((....))))..))))............... ( -30.90, z-score = -2.33, R) >droSec1.super_0 8770749 103 - 21120651 -----------------AACCUGCUCGGGAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUGUACCUCCUUCGGAAAUAUUUCCUGUCGAAUUACCACUUCCGGA -----------------........((((((....((((...(((..(((......(((((....)))))......)))..)))...))))....))))))........((......)). ( -26.40, z-score = -1.37, R) >droSim1.chr3L 16025235 103 - 22553184 -----------------AACCUGCUCGGGAAUAGGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUUUACCUCCUUCGGAAAUAUUUCCUGUCGAAUUACCACUUCCGGA -----------------.........(((((((..((((...(((...........(((((....)))))...........)))...)))).)))))))..........((......)). ( -23.15, z-score = 0.01, R) >consensus _________________AACCUGCUCGGGAAUAAGUUCCUCUGGAUUUACUACCAAUGGACUUGCGUCCAGUCGAUUUACCUCCUUCGGAAUUAUUUCCUGUCGAAUUACCACUUCCGGA ..........................((((((.((((((...(((...........(((((....)))))...........)))...))))))))))))..................... (-17.89 = -18.69 + 0.80)

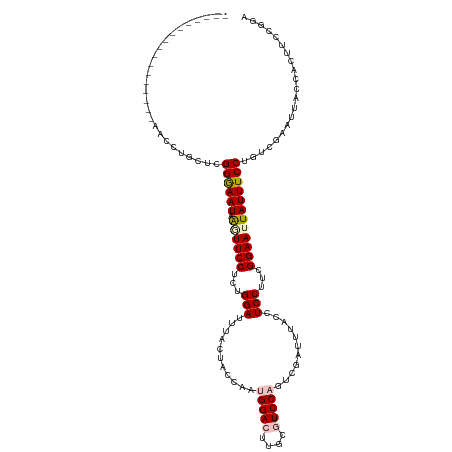
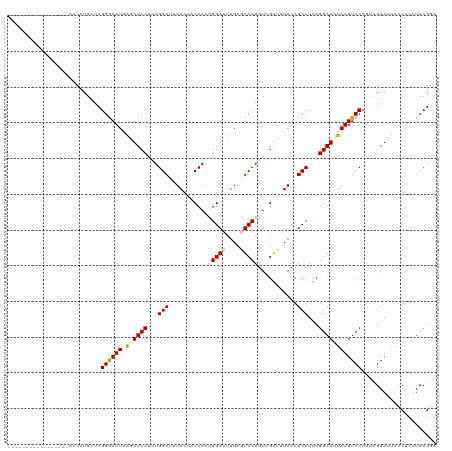
| Location | 16,698,421 – 16,698,520 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Shannon entropy | 0.37348 |
| G+C content | 0.50136 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -19.49 |
| Energy contribution | -20.57 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16698421 99 + 24543557 CGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUUCCGAGCAAGUUAUACCGGUGC------UGGUAGUUCCGGAUCUGA--------------- ....(((..((((((((.(((((....)))))))))).)))..)))...((((((.....((.((((.(......)..)))------))).))))))........--------------- ( -34.70, z-score = -2.80, R) >droEre2.scaffold_4784 8626352 120 - 25762168 CGAAGGAGGUAAAUCAACUGGAUGCAAUUCCGGUGGAAGUAAUUCCAAAGGAACUAAUUCCCGGACAGGUUAUACCGGUGGAUGUAAUGGUAAUGCCGGUGCUGGUAGUUCCGGAGCUGG ....((((.((..((.((((((......)))))).))..)).))))...(((((((....((((((.(((..(((((..........)))))..))).)).)))))))))))........ ( -37.40, z-score = -1.31, R) >droYak2.chr3L 6821013 120 - 24197627 CGAAGGAGGUAAACCGACUGGAUGCAAUUCCGGUGGAUGUAAUUCCAGAGGAAUUAAUUCCCGAGCAGGUAAAACCGGUGGAGGUAAUGGUAAUACCGGUGCUGGUAGUUCAGGAGCUGG ...((..((....))..))....((((((((..((((......))))..))))))...(((.((((.(.((..(((((((.............)))))))..)).).)))).)))))... ( -36.62, z-score = -1.42, R) >droSec1.super_0 8770781 99 + 21120651 CGAAGGAGGUACAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGGUUAUACCGGUGC------UGGCAGUUCCGGAUCUGA--------------- ....(((..((((((((.(((((....)))))))))).)))..)))...((((((.....((.((((((.....))..)))------))).))))))........--------------- ( -35.20, z-score = -2.29, R) >droSim1.chr3L 16025267 99 + 22553184 CGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACCUAUUCCCGAGCAGGUUAUACCGGUGC------UGGCAGUUCCGGAUCUGA--------------- ....(((..((.(((((.(((((....))))))))))..))..))).(.((((....)))))...(((((....((((.((------.....)).))))))))).--------------- ( -31.30, z-score = -1.12, R) >consensus CGAAGGAGGUAAAUCGACUGGACGCAAGUCCAUUGGUAGUAAAUCCAGAGGAACUUAUUCCCGAGCAGGUUAUACCGGUGC______UGGUAGUUCCGGAUCUGA_______________ ((..(((..((..((((.(((((....)))))))))...))..)))...((((....))))))..(((((....((((.................)))))))))................ (-19.49 = -20.57 + 1.08)

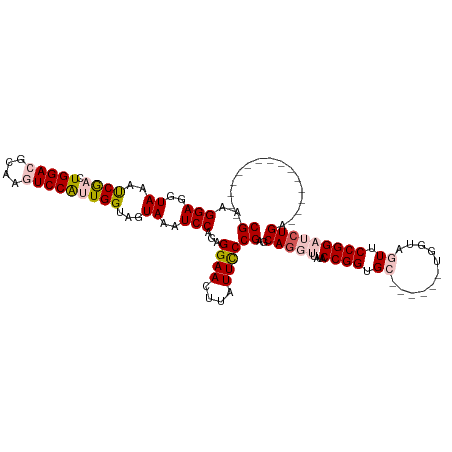
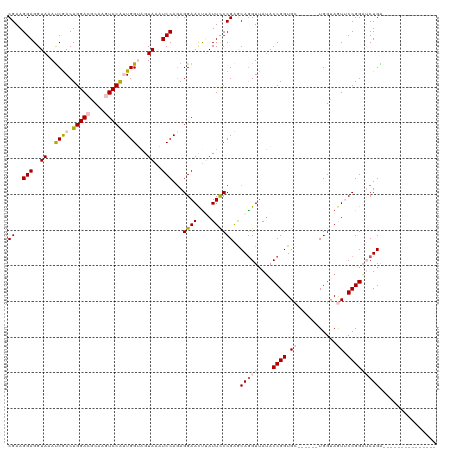
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:59 2011