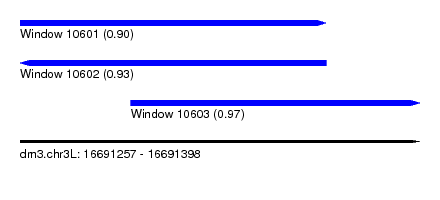
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,691,257 – 16,691,398 |
| Length | 141 |
| Max. P | 0.973173 |

| Location | 16,691,257 – 16,691,365 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.38668 |
| G+C content | 0.52681 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -22.91 |
| Energy contribution | -22.42 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16691257 108 + 24543557 -CUUUUAGUCCAGCAUGGACCACUGUUGGUUGGCCAAGUGGAAUCGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCUGU---GGAGCACUCUUC -.......(((.....)))(((((..(((....))))))))....(((((((((((.((((((..(....)..)))))).)))).)))..((((...---.))))))))... ( -38.20, z-score = -1.79, R) >droAna3.scaffold_13337 9383256 98 - 23293914 CUUCUAUGUCUUCCUCAGACCACUGUACU--------------CCAAGUGGAAUGGAGUUGUCCCCACCAGAAGACAACACUGUUUCUGUUCUCAGUUCCAGUACCAGUACC ......(((((((....(((.(((.((.(--------------((....))).)).))).))).......)))))))..((((...(((....)))...))))......... ( -23.70, z-score = -0.73, R) >droEre2.scaffold_4784 8619403 104 - 25762168 -CUUUUAGACCAGCAUGGACCACUGUUGC----CGAAGUGGAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCUGAGCUCGGC---GGAACCCUCUUC -......(.(((((.(((((((((....(----(.....)).....))))).((((.((((((..(....)..)))))).)))).)))).))).)))---............ ( -28.60, z-score = -0.20, R) >droYak2.chr3L 6813962 104 - 24197627 -CUUUUAGUCCAGCAUGGACCACUGUUGC----CCAAGUGGAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGA---GAAACCCUCUUC -.......((((((.(((((((((.....----((....)).....))))).((((.((((((..(....)..)))))).)))).)))).))).)))---............ ( -32.90, z-score = -2.12, R) >droSec1.super_0 8763791 104 + 21120651 -CUUUUUGUCCAGCAUGGACCACUGUUGC----CCAAGUGGAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU---GGAGUCCACUUC -.......((((((.....(((((.....----...)))))....(((((((((((.((((((..(....)..)))))).)))).)))..)))))))---)))......... ( -38.50, z-score = -2.74, R) >droSim1.chr3L 16018255 104 + 22553184 -CUUUUUGUCCAGCAUGGACCACUGUUGC----CCAAGUGGAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU---GGAGUCCACUUC -.......((((((.....(((((.....----...)))))....(((((((((((.((((((..(....)..)))))).)))).)))..)))))))---)))......... ( -38.50, z-score = -2.74, R) >consensus _CUUUUAGUCCAGCAUGGACCACUGUUGC____CCAAGUGGAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGU___GGAACCCUCUUC ...........(((.(((((((((......................))))).((((.((((((..(....)..)))))).)))).)))).)))................... (-22.91 = -22.42 + -0.50)

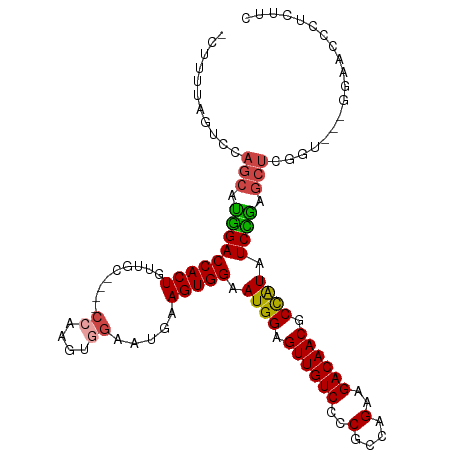
| Location | 16,691,257 – 16,691,365 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.38668 |
| G+C content | 0.52681 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16691257 108 - 24543557 GAAGAGUGCUCC---ACAGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCGAUUCCACUUGGCCAACCAACAGUGGUCCAUGCUGGACUAAAAG- ...((((((((.---...))))..(((.((((.(((((((((....))))))))).)))))))))))...(((((.((((((........))).))).).)))).......- ( -40.40, z-score = -2.75, R) >droAna3.scaffold_13337 9383256 98 + 23293914 GGUACUGGUACUGGAACUGAGAACAGAAACAGUGUUGUCUUCUGGUGGGGACAACUCCAUUCCACUUGG--------------AGUACAGUGGUCUGAGGAAGACAUAGAAG ..(((((.(((((...(((....)))...))))).(((((((....)))))))((((((.......)))--------------))).)))))((((.....))))....... ( -30.90, z-score = -0.82, R) >droEre2.scaffold_4784 8619403 104 + 25762168 GAAGAGGGUUCC---GCCGAGCUCAGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUUCAUUCCACUUCG----GCAACAGUGGUCCAUGCUGGUCUAAAAG- ((((.((((((.---...)))))).(..((((.(((((((((....))))))))).))))..).))))...(((((..(----....)))))).((.....))........- ( -34.10, z-score = -1.23, R) >droYak2.chr3L 6813962 104 + 24197627 GAAGAGGGUUUC---UCCGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUUCAUUCCACUUGG----GCAACAGUGGUCCAUGCUGGACUAAAAG- ((((.((((((.---...))))))(((.((((.(((((((((....))))))))).))))))).))))..(((((.(((----((.......))))).).)))).......- ( -39.70, z-score = -2.67, R) >droSec1.super_0 8763791 104 - 21120651 GAAGUGGACUCC---AGCGAGCUUGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUUCCACUUGG----GCAACAGUGGUCCAUGCUGGACAAAAAG- .........(((---((((((..((((.((((.(((((((((....))))))))).)))))))))))....(((((..(----....)))))).....)))))).......- ( -40.30, z-score = -2.56, R) >droSim1.chr3L 16018255 104 - 22553184 GAAGUGGACUCC---AGCGAGCUUGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUUCCACUUGG----GCAACAGUGGUCCAUGCUGGACAAAAAG- .........(((---((((((..((((.((((.(((((((((....))))))))).)))))))))))....(((((..(----....)))))).....)))))).......- ( -40.30, z-score = -2.56, R) >consensus GAAGAGGGCUCC___ACCGAGCUCGGAUAUGGCGUUGUCUUCUGGCGGGGACAACUCCAUUCCACUCCAUUCCACUUGG____GCAACAGUGGUCCAUGCUGGACAAAAAG_ ..................(((...(((.((((.(((((((((....))))))))).))))))).))).........................((((.....))))....... (-22.84 = -23.15 + 0.31)
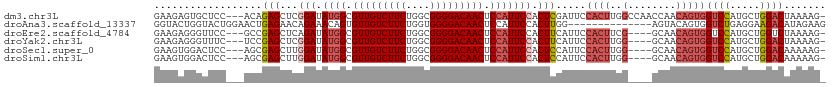
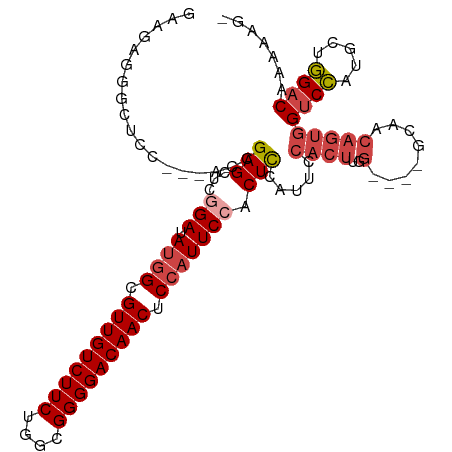
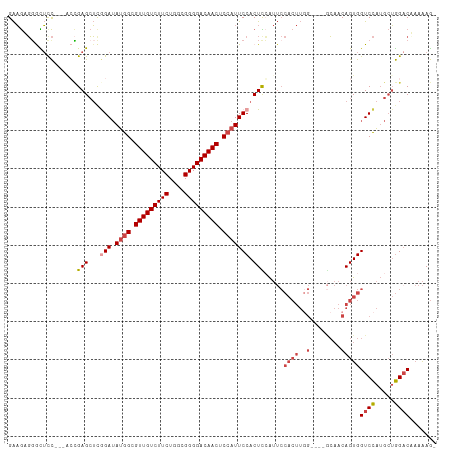
| Location | 16,691,296 – 16,691,398 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Shannon entropy | 0.39809 |
| G+C content | 0.55271 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.47 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.973173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16691296 102 + 24543557 GAAUCGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCUGU---GGAGCACUCUUCACCGCCCCUUCUUCCCCCCUUCA-CUUAGCGCAC (((..(((((((((((.((((((..(....)..)))))).)))).)))..((((...---.)))))))))))..(((..................-....)))... ( -26.95, z-score = -1.25, R) >droAna3.scaffold_13337 9383288 94 - 23293914 ------AGUGGAAUGGAGUUGUCCCCACCAGAAGACAACACUGUUUCUGUUCUCAGUUCCAGUACCAGUACCAGCCCCA------UUCCUCUUCAGUUUGGAGCAC ------...(((((((.((((((..(....)..))))))((((...(((....)))...)))).............)))------)))).((((.....))))... ( -25.00, z-score = -1.44, R) >droEre2.scaffold_4784 8619438 92 - 25762168 GAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCUGAGCUCGGC---GGAACCCUCUUCUCCGC----------CCCCUUCA-CUUAGAGCAC ...(((((.((.((((.((((((..(....)..)))))).))))..........(((---(((.........)))))----------))))))))-.......... ( -29.90, z-score = -2.40, R) >droYak2.chr3L 6813997 92 - 24197627 GAAUGAAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGA---GAAACCCUCUUCUCCGC----------CCCCUUCA-CUUAGAGCAC ...(((((((((((((.((((((..(....)..)))))).)))).))))....((((---(((......))))))).----------...)))))-.......... ( -29.90, z-score = -3.35, R) >droSec1.super_0 8763826 93 + 21120651 GAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU---GGAGUCCACUUCACCCCG---------CUUCUUCA-CUUAGCGCAC ...(((((((((((((.((((((..(....)..)))))).)))).((((.(....))---))).)))))))))...((---------((......-...))))... ( -32.20, z-score = -2.94, R) >droSim1.chr3L 16018290 93 + 22553184 GAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCAAGCUCGCU---GGAGUCCACUUCACCCCC---------CUUCUUCA-CUUAGCGCAC ...(((((((((((((.((((((..(....)..)))))).)))).((((.(....))---))).))))))))).....---------........-.......... ( -29.20, z-score = -2.40, R) >consensus GAAUGGAGUGGAAUGGAGUUGUCCCCGCCAGAAGACAACGCCAUAUCCGAGCUCGGU___GGAACCCUCUUCACCCCC_________CCCCUUCA_CUUAGAGCAC ...(((((((((((((.((((((..(....)..)))))).)))).))))...........((((....))))..................)))))........... (-17.91 = -17.47 + -0.44)
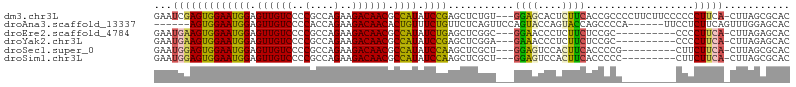
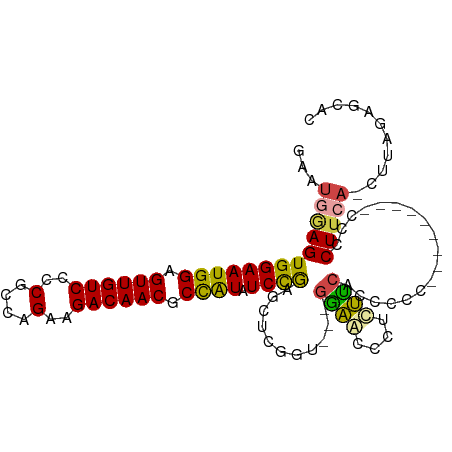
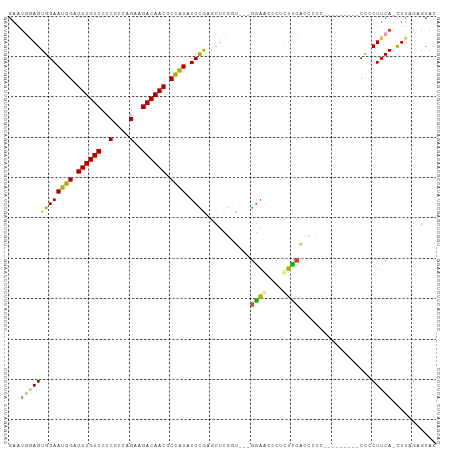
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:53 2011