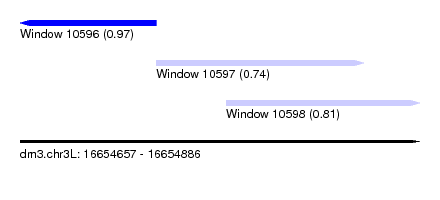
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,654,657 – 16,654,886 |
| Length | 229 |
| Max. P | 0.968614 |

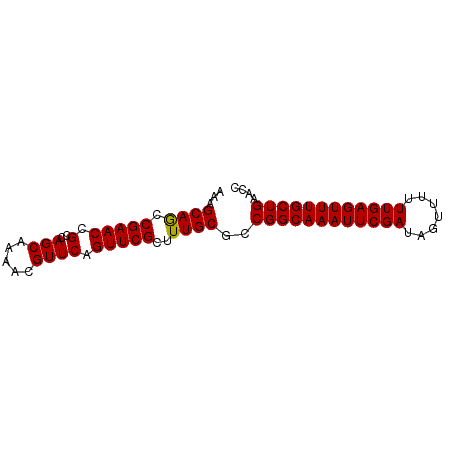
| Location | 16,654,657 – 16,654,735 |
|---|---|
| Length | 78 |
| Sequences | 5 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 97.69 |
| Shannon entropy | 0.04021 |
| G+C content | 0.48205 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.74 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 1.01 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16654657 78 - 24543557 AAAGCAGCCGAACCGCCAGCUAAACGUUCAGUUCGCUUUGCGCCGGCAAAUUCGAUAGUUUUUUGAGUUUGCUGAACC ((((((((.((((............)))).))).)))))....((((((((((((.......)))))))))))).... ( -22.70, z-score = -1.57, R) >droSim1.chr3L 15985686 78 - 22553184 AAAGCAGCCGAACCGCCAGCAAAACGUUCAGUUCGCUUUGCGCCGGCAAAUUCGAUAGUUUUUUGAGUUUGCUGAACC ((((((((.((((............)))).))).)))))....((((((((((((.......)))))))))))).... ( -22.70, z-score = -1.54, R) >droSec1.super_0 8732469 78 - 21120651 AAAGCAGCCGAACCGCCAGCAAAACGUUCAGUUCGCUUUGCGCCGGCAAAUUCGAUAGUUUUUUGAGUUUGCUGAACC ((((((((.((((............)))).))).)))))....((((((((((((.......)))))))))))).... ( -22.70, z-score = -1.54, R) >droYak2.chr3L 6781686 78 + 24197627 AAAGCAACCGAACCGCCAGCAAAACGUUCAGUUCGCUUUGCGCCGGCAAAUUCGAUAGAUUUUUGAGUUUGCUGAACC ...((((.(((((.(..(((.....)))).)))))..))))..((((((((((((.......)))))))))))).... ( -23.00, z-score = -2.22, R) >droEre2.scaffold_4784 8588329 78 + 25762168 AAAGCAGCCGAACCGCCAGCAAAACGUUCAGUUCGCUUUGCACCGGCAAAUUCGAUAGAUUUUUGAGUUUGCUGAACC ((((((((.((((............)))).))).)))))....((((((((((((.......)))))))))))).... ( -22.70, z-score = -2.28, R) >consensus AAAGCAGCCGAACCGCCAGCAAAACGUUCAGUUCGCUUUGCGCCGGCAAAUUCGAUAGUUUUUUGAGUUUGCUGAACC ...((((.(((((.(..(((.....)))).)))))..))))..((((((((((((.......)))))))))))).... (-22.90 = -22.74 + -0.16)

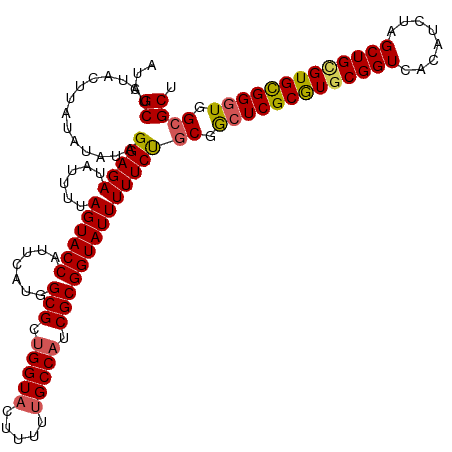
| Location | 16,654,735 – 16,654,854 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Shannon entropy | 0.12948 |
| G+C content | 0.47248 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -35.69 |
| Energy contribution | -35.65 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16654735 119 + 24543557 AUCGCUAUACUUAUAUAUAGGAGAUAUUUUAGUACCAAUCAUGGCGCUGGUACUUUU-GCCAUCGCGGUAUUUUUCUGCGGCUCGCGUGCGGUCACAUCUAGCUGCGUGCGGGUGGAGCU ...(((...........((((((.(((..(((..(((....)))..)))))))))))-)((..(((((.......)))))(((((((((((((........)))))))))))))))))). ( -37.20, z-score = -0.48, R) >droEre2.scaffold_4784 8588407 120 - 25762168 AUCGCUAUUCUUAUAUAUAGGAGAUAUUUUAGUACCUUUUAUGGCGUUGGUACUUUUUGCCAUCGCGGUAUUUUUCCGCGAAUCGCAUGCGGUCACACUUAGCUGUGUGUGGGUGGCGCU ...((..((((((....)))))).......((((((............))))))....((((((((((.......))))....((((((((((........)))))))))))))))))). ( -38.70, z-score = -2.40, R) >droYak2.chr3L 6781764 120 - 24197627 AUCGCUAUACUUAUAUACCGGAGAUAUUUUAGUACCUUUUAUGGCGUUGGUACUUUUUGCCAUCGCGGUAUUUUUCUGCGACUCGCAUGCGGUCACAUUUAGCUGAGUGUGGGUGGCGCU ...((.........((((((((........((((((............))))))........)).))))))......((.((((((((.((((........)))).)))))))).)))). ( -36.39, z-score = -1.62, R) >droSec1.super_0 8732547 120 + 21120651 AUCGCUAUACUUAUAUAUAGGAGAUAUUUUAGUACCAUUCAUGGCGCUGGUACUUUUUGCCAUCGCGGUAUUUUUCUGCGGCUCGCGUGCGGUCACAUCUAGCUGCGUGCGGGUGGCGCU (((.(((((......)))))..))).....(((((((..........)))))))....(((..(((((.......)))))(((((((((((((........))))))))))))))))... ( -40.00, z-score = -1.20, R) >droSim1.chr3L 15985764 120 + 22553184 AUCGCUAUACUUAUAUAUAGGAGAUAUUUUAGUACCAUUCAUGGCGCUGGUACUUUUUGCCCUCGCGGUAUUUUUCUGCGGCUCGCGUGCGGUCACAUCUAGCUGUGUGCGGGUGGCGCU (((.(((((......)))))..))).....(((((((..........)))))))....(((..(((((.......)))))(((((((..((((........))))..))))))))))... ( -39.10, z-score = -1.00, R) >consensus AUCGCUAUACUUAUAUAUAGGAGAUAUUUUAGUACCAUUCAUGGCGCUGGUACUUUUUGCCAUCGCGGUAUUUUUCUGCGGCUCGCGUGCGGUCACAUCUAGCUGCGUGCGGGUGGCGCU ...((..............(((((......((((((.......(((.(((((.....))))).))))))))))))))((.(((((((((((((........))))))))))))).)))). (-35.69 = -35.65 + -0.04)

| Location | 16,654,775 – 16,654,886 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.64 |
| Shannon entropy | 0.20864 |
| G+C content | 0.53906 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -36.55 |
| Energy contribution | -35.80 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16654775 111 + 24543557 AUGGCGCUGGUACUUUU-GCCAUCGCGGUAUUUUUCUGCGGCUCGCGUGCGGUCACAUCUAGCUGCGUGCGGGUGGAGCUCUAUGCACACUGGUUCCACUCCUAUUGGGUAC-------- ((((((..(....)..)-)))))(((((.......)))))(((((((((((((........)))))))))((((((((((...........)))))))))).....))))..-------- ( -39.10, z-score = -0.80, R) >droEre2.scaffold_4784 8588447 120 - 25762168 AUGGCGUUGGUACUUUUUGCCAUCGCGGUAUUUUUCCGCGAAUCGCAUGCGGUCACACUUAGCUGUGUGUGGGUGGCGCUCUCUGCACACUGGUUCCACUCUUAUGAGACACAAAACAGU .(((..(..((.......((((((((((.......))))....((((((((((........))))))))))))))))((.....))..))..)..)))(((....)))............ ( -39.90, z-score = -1.51, R) >droYak2.chr3L 6781804 112 - 24197627 AUGGCGUUGGUACUUUUUGCCAUCGCGGUAUUUUUCUGCGACUCGCAUGCGGUCACAUUUAGCUGAGUGUGGGUGGCGCUCUCUGCACAUUGGUUCCACUAUUAUUGCGCAC-------- ...(((((((.(((....((((((((((.......))))))(((((((.((((........)))).)))))))))))((.....)).....))).)))........))))..-------- ( -35.30, z-score = -0.99, R) >droSec1.super_0 8732587 112 + 21120651 AUGGCGCUGGUACUUUUUGCCAUCGCGGUAUUUUUCUGCGGCUCGCGUGCGGUCACAUCUAGCUGCGUGCGGGUGGCGCUCUCUGCACACUGGUUCCACUUUUAUUGGGCAC-------- .(((.((..((.......(((..(((((.......)))))(((((((((((((........))))))))))))))))((.....))..))..)).)))..............-------- ( -41.00, z-score = -1.54, R) >consensus AUGGCGCUGGUACUUUUUGCCAUCGCGGUAUUUUUCUGCGACUCGCAUGCGGUCACAUCUAGCUGCGUGCGGGUGGCGCUCUCUGCACACUGGUUCCACUCUUAUUGGGCAC________ .(((..(..((.......(((..(((((.......)))))(((((((((((((........))))))))))))))))((.....))..))..)..)))...................... (-36.55 = -35.80 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:49 2011