
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,650,862 – 16,651,004 |
| Length | 142 |
| Max. P | 0.998752 |

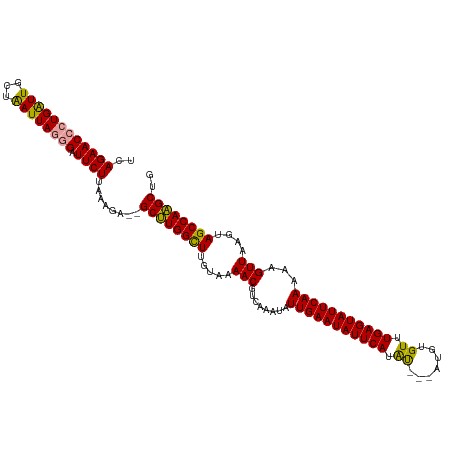
| Location | 16,650,862 – 16,650,976 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.17 |
| Shannon entropy | 0.13343 |
| G+C content | 0.34602 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -31.11 |
| Energy contribution | -30.23 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.998752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16650862 114 + 24543557 UCAGAACCGUGGUUGCUAAUUAGAGAUUCUUAAACAUGGCAUGGUUUGGAAAACGUCAAAUAUUGAAUAUUCAUGC---AUGUGUUUGAGUAUUCAAGAAGUUAAGUAGCCAUGCUG .(((...((((((((((((((...((..(((((((((((((((((((((......))))))..........)))))---.))))))))))...))....)))).))))))))))))) ( -30.70, z-score = -2.47, R) >droSim1.chr3L 15981705 115 + 22553184 UCAGAACCCUGAUUGCUAAUUAGGGAUUCUUAAAGA--GCUUGGCUUGUAAAACGUCAAAUAUUGAAUAUUCAUAUGCUAUGUGUUUGAGUAUUCAAAAAGUUAAGUAGCCAAGCUG ..(((((((((((.....))))))).)))).....(--((((((((.....(((........(((((((((((.((((...)))).)))))))))))...)))....))))))))). ( -36.90, z-score = -5.03, R) >droSec1.super_0 8728772 112 + 21120651 UCAGAACCCUGAUUGCUGAUUAGGGAUUCUUAAAGA--GCUUGGCUUGUAAAACGUCAAAUAUUGAAUAUUCAUAU---AUGUGUUUGAGUAUUCAAAAAGUUAAGUAGCCAAGCUG ..((((((((((((...)))))))).)))).....(--((((((((.....(((........(((((((((((.((---....)).)))))))))))...)))....))))))))). ( -35.70, z-score = -4.99, R) >consensus UCAGAACCCUGAUUGCUAAUUAGGGAUUCUUAAAGA__GCUUGGCUUGUAAAACGUCAAAUAUUGAAUAUUCAUAU___AUGUGUUUGAGUAUUCAAAAAGUUAAGUAGCCAAGCUG ..((((((((((((...)))))))).))))........((((((((.....(((........(((((((((((.((.......)).)))))))))))...)))....)))))))).. (-31.11 = -30.23 + -0.88)


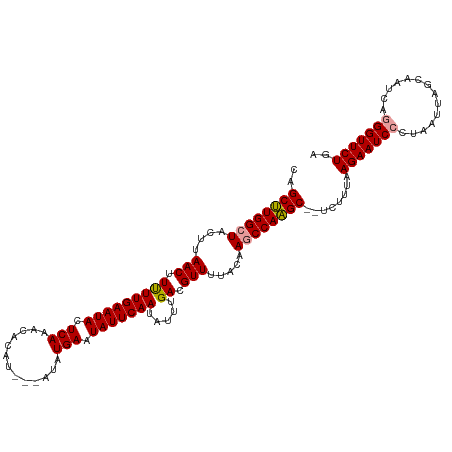
| Location | 16,650,862 – 16,650,976 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.17 |
| Shannon entropy | 0.13343 |
| G+C content | 0.34602 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.06 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16650862 114 - 24543557 CAGCAUGGCUACUUAACUUCUUGAAUACUCAAACACAU---GCAUGAAUAUUCAAUAUUUGACGUUUUCCAAACCAUGCCAUGUUUAAGAAUCUCUAAUUAGCAACCACGGUUCUGA .((((((((...........(((((((.(((..(....---)..))).)))))))..((((........))))....)))))))).............((((.(((....))))))) ( -20.90, z-score = -1.67, R) >droSim1.chr3L 15981705 115 - 22553184 CAGCUUGGCUACUUAACUUUUUGAAUACUCAAACACAUAGCAUAUGAAUAUUCAAUAUUUGACGUUUUACAAGCCAAGC--UCUUUAAGAAUCCCUAAUUAGCAAUCAGGGUUCUGA .(((((((((....(((((.(((((((.(((.............))).))))))).....)).))).....))))))))--).....((((.((((.((.....)).)))))))).. ( -26.52, z-score = -3.04, R) >droSec1.super_0 8728772 112 - 21120651 CAGCUUGGCUACUUAACUUUUUGAAUACUCAAACACAU---AUAUGAAUAUUCAAUAUUUGACGUUUUACAAGCCAAGC--UCUUUAAGAAUCCCUAAUCAGCAAUCAGGGUUCUGA .(((((((((....(((((.(((((((.(((.......---...))).))))))).....)).))).....))))))))--).....((((.((((.((.....)).)))))))).. ( -26.70, z-score = -3.69, R) >consensus CAGCUUGGCUACUUAACUUUUUGAAUACUCAAACACAU___AUAUGAAUAUUCAAUAUUUGACGUUUUACAAGCCAAGC__UCUUUAAGAAUCCCUAAUUAGCAAUCAGGGUUCUGA ..((((((((....(((.(((((((((.(((.............))).))))))).....)).))).....))))))))........(((((((..............))))))).. (-21.06 = -21.06 + 0.00)


| Location | 16,650,902 – 16,651,004 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Shannon entropy | 0.20976 |
| G+C content | 0.35685 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -20.04 |
| Energy contribution | -18.93 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16650902 102 + 24543557 AUGGUUUGGAAAACGUCAAAUAUUGAAUAUUCAUGC---AUGUGUUUGAGUAUUCAAGAAGUUAAGUAGCCAUGCUGAGCA---AGCGGAAAUCAAUCC---------GUUUUAAUU ..(((.(((...((........(((((((((((.((---....)).)))))))))))........))..))).)))....(---((((((......)))---------))))..... ( -26.59, z-score = -2.72, R) >droSim1.chr3L 15981743 117 + 22553184 UUGGCUUGUAAAACGUCAAAUAUUGAAUAUUCAUAUGCUAUGUGUUUGAGUAUUCAAAAAGUUAAGUAGCCAAGCUGAGCAGCAAGCGGAAGUCAACCCUUGGGCACCGUUUUAAUU ((((((.....(((........(((((((((((.((((...)))).)))))))))))...)))....))))))(((....)))((((((..(((........))).))))))..... ( -28.60, z-score = -1.35, R) >droSec1.super_0 8728810 111 + 21120651 UUGGCUUGUAAAACGUCAAAUAUUGAAUAUUCAUAU---AUGUGUUUGAGUAUUCAAAAAGUUAAGUAGCCAAGCUGAGUG---AGCGGAAGUCAACCCUUGGGCACAGUUUUAAUU ((((((.....(((........(((((((((((.((---....)).)))))))))))...)))....))))))((((..((---(.(....)))).((....))..))))....... ( -25.00, z-score = -1.36, R) >consensus UUGGCUUGUAAAACGUCAAAUAUUGAAUAUUCAUAU___AUGUGUUUGAGUAUUCAAAAAGUUAAGUAGCCAAGCUGAGCA___AGCGGAAGUCAACCCUUGGGCAC_GUUUUAAUU ((((((.....(((........(((((((((((.((.......)).)))))))))))...)))....))))))(((........)))((........)).................. (-20.04 = -18.93 + -1.10)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:47 2011