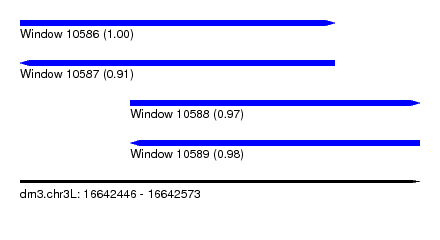
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,642,446 – 16,642,573 |
| Length | 127 |
| Max. P | 0.998710 |

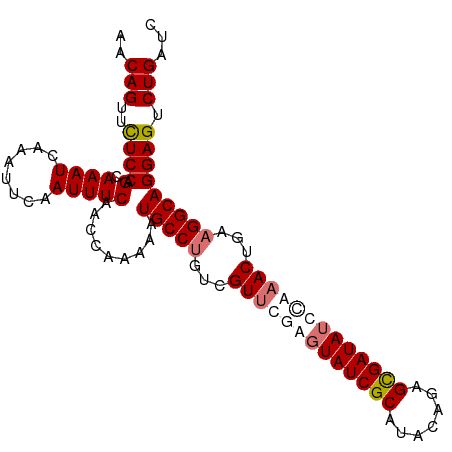
| Location | 16,642,446 – 16,642,546 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.50 |
| Shannon entropy | 0.18547 |
| G+C content | 0.39750 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -25.04 |
| Energy contribution | -26.85 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.998710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
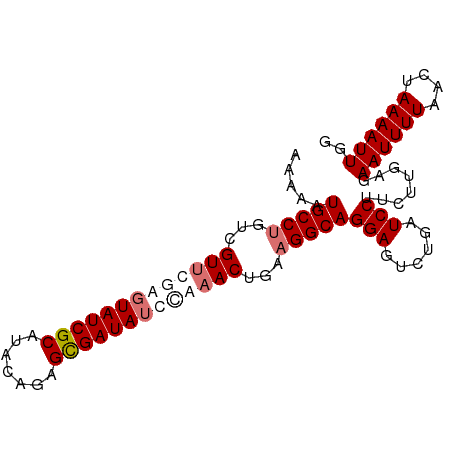
>dm3.chr3L 16642446 100 + 24543557 AACAGUUCUCCAGUAAAUCAAAUUCAAUUUCAACCAAAAAUGCCUGUCGUUCAAGUAUCGCAAACUGUGCGAUAUCCAAACUGAAGGCAGGAGUCUGAUC ..(((..((((.(.((((........))))).........(((((...(((...((((((((.....))))))))...)))...))))))))).)))... ( -26.10, z-score = -3.96, R) >droSim1.chr3L 15973226 100 + 22553184 AACAGUUCUCCAGCAAAUCAAAUUCAAUUUCAACCAAAAAUGCCUGUCGUUCGAGUAUCGCAUACUGUGUGAUAUCCAAACUGAAGGCAGGAGUCUGAUC ..(((..((((.(.((((........))))).........(((((...(((.(..((((((((...))))))))..).)))...))))))))).)))... ( -26.20, z-score = -3.08, R) >droYak2.chr3L 6769365 100 - 24197627 AACAGUUUUCCAGCAAAUCAAAUUCUAUUUCAACCAAGAAUGCCCGACGUUUGAUUAUCGCAUACAAAGCGAUAUUGAAACUUUAGGCAGGAGUCUGAUC ..(((..((((..........(((((..........)))))(((..(.((((.(.((((((.......)))))).).)))).)..))).)))).)))... ( -26.00, z-score = -3.58, R) >droEre2.scaffold_4784 8576260 100 - 25762168 AACAGUUCUCCAGCAAAUCAAAUUCAAUUUCAACCAAGAUUGCCUGUCGUUUGAGUAUCGCAUACAGAGCGAUAUUUACACUAAAGGCAGGAGUCUGAUC ..(((..((((..................((......)).(((((...((.((((((((((.......)))))))))).))...))))))))).)))... ( -29.30, z-score = -4.33, R) >consensus AACAGUUCUCCAGCAAAUCAAAUUCAAUUUCAACCAAAAAUGCCUGUCGUUCGAGUAUCGCAUACAGAGCGAUAUCCAAACUGAAGGCAGGAGUCUGAUC ..(((..((((.(.((((........))))).........(((((...(((((((((((((.......)))))))))))))...))))))))).)))... (-25.04 = -26.85 + 1.81)

| Location | 16,642,446 – 16,642,546 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.50 |
| Shannon entropy | 0.18547 |
| G+C content | 0.39750 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -23.02 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16642446 100 - 24543557 GAUCAGACUCCUGCCUUCAGUUUGGAUAUCGCACAGUUUGCGAUACUUGAACGACAGGCAUUUUUGGUUGAAAUUGAAUUUGAUUUACUGGAGAACUGUU ...(((.(((((((((...(((..(.(((((((.....))))))).)..)))...))))).....(((..((......))..)))....))))..))).. ( -28.40, z-score = -2.78, R) >droSim1.chr3L 15973226 100 - 22553184 GAUCAGACUCCUGCCUUCAGUUUGGAUAUCACACAGUAUGCGAUACUCGAACGACAGGCAUUUUUGGUUGAAAUUGAAUUUGAUUUGCUGGAGAACUGUU ...(((.(((((((((...(((((..((((.((.....)).))))..)))))...))))).....(((..(((((......))))))))))))..))).. ( -25.60, z-score = -1.47, R) >droYak2.chr3L 6769365 100 + 24197627 GAUCAGACUCCUGCCUAAAGUUUCAAUAUCGCUUUGUAUGCGAUAAUCAAACGUCGGGCAUUCUUGGUUGAAAUAGAAUUUGAUUUGCUGGAAAACUGUU (((((((.((.(((((...((((.(.((((((.......)))))).).))))...)))))(((......)))...)).)))))))............... ( -22.80, z-score = -1.18, R) >droEre2.scaffold_4784 8576260 100 + 25762168 GAUCAGACUCCUGCCUUUAGUGUAAAUAUCGCUCUGUAUGCGAUACUCAAACGACAGGCAAUCUUGGUUGAAAUUGAAUUUGAUUUGCUGGAGAACUGUU ...(((.(((((((((...((.....((((((.......)))))).....))...))))).....(((..(((((......))))))))))))..))).. ( -23.40, z-score = -1.20, R) >consensus GAUCAGACUCCUGCCUUCAGUUUGAAUAUCGCACAGUAUGCGAUACUCAAACGACAGGCAUUCUUGGUUGAAAUUGAAUUUGAUUUGCUGGAGAACUGUU ...(((.(((((((((...((((((.((((((.......)))))).))))))...))))).....(((..((......))..)))....))))..))).. (-23.02 = -23.52 + 0.50)

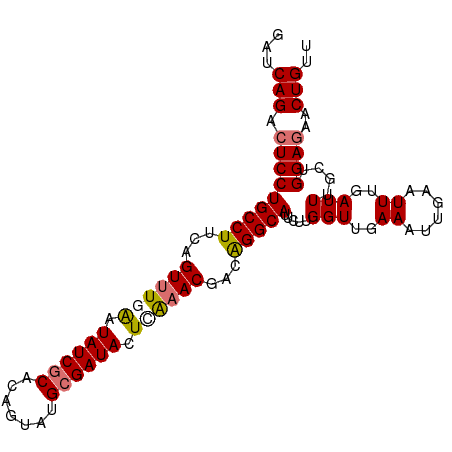
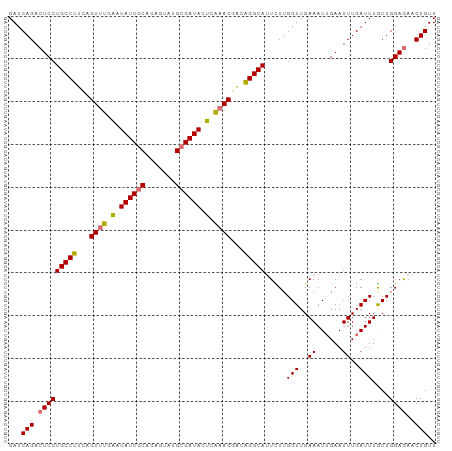
| Location | 16,642,481 – 16,642,573 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.13 |
| Shannon entropy | 0.17514 |
| G+C content | 0.38315 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -20.77 |
| Energy contribution | -22.77 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.974941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16642481 92 + 24543557 AAAAAUGCCUGUCGUUCAAGUAUCGCAAACUGUGCGAUAUCCAAACUGAAGGCAGGAGUCUGAUCCUUCUUGAGAAUUUUAACUAAAAUUGG .....(((((...(((...((((((((.....))))))))...)))...)))))(((......)))........((((((....)))))).. ( -21.40, z-score = -1.82, R) >droSim1.chr3L 15973261 92 + 22553184 AAAAAUGCCUGUCGUUCGAGUAUCGCAUACUGUGUGAUAUCCAAACUGAAGGCAGGAGUCUGAUCCUUCUUGAGAAUUUUAACUAAAAUUGG .....(((((...(((.(..((((((((...))))))))..).)))...)))))(((......)))........((((((....)))))).. ( -21.50, z-score = -1.69, R) >droYak2.chr3L 6769400 92 - 24197627 AAGAAUGCCCGACGUUUGAUUAUCGCAUACAAAGCGAUAUUGAAACUUUAGGCAGGAGUCUGAUCCUUCUUGAGAAUUUUAACUAAAAUUGG (((((((((..(.((((.(.((((((.......)))))).).)))).)..))))(((......))))))))...((((((....)))))).. ( -23.70, z-score = -2.80, R) >droEre2.scaffold_4784 8576295 92 - 25762168 AAGAUUGCCUGUCGUUUGAGUAUCGCAUACAGAGCGAUAUUUACACUAAAGGCAGGAGUCUGAUCCUUCUUGAGAAUUUUAACUAAAAUUGG ((((.(((((...((.((((((((((.......)))))))))).))...)))))(((......))).))))...((((((....)))))).. ( -25.10, z-score = -2.51, R) >consensus AAAAAUGCCUGUCGUUCGAGUAUCGCAUACAGAGCGAUAUCCAAACUGAAGGCAGGAGUCUGAUCCUUCUUGAGAAUUUUAACUAAAAUUGG .....(((((...(((((((((((((.......)))))))))))))...)))))(((......)))........((((((....)))))).. (-20.77 = -22.77 + 2.00)

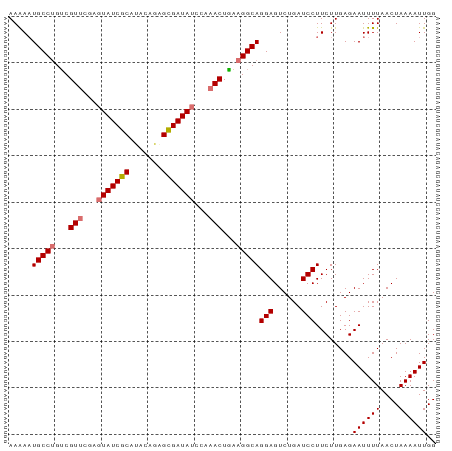
| Location | 16,642,481 – 16,642,573 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.13 |
| Shannon entropy | 0.17514 |
| G+C content | 0.38315 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -19.90 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16642481 92 - 24543557 CCAAUUUUAGUUAAAAUUCUCAAGAAGGAUCAGACUCCUGCCUUCAGUUUGGAUAUCGCACAGUUUGCGAUACUUGAACGACAGGCAUUUUU .........................((((......))))((((...(((..(.(((((((.....))))))).)..)))...))))...... ( -23.70, z-score = -2.60, R) >droSim1.chr3L 15973261 92 - 22553184 CCAAUUUUAGUUAAAAUUCUCAAGAAGGAUCAGACUCCUGCCUUCAGUUUGGAUAUCACACAGUAUGCGAUACUCGAACGACAGGCAUUUUU .........................((((......))))((((...(((((..((((.((.....)).))))..)))))...))))...... ( -20.60, z-score = -2.02, R) >droYak2.chr3L 6769400 92 + 24197627 CCAAUUUUAGUUAAAAUUCUCAAGAAGGAUCAGACUCCUGCCUAAAGUUUCAAUAUCGCUUUGUAUGCGAUAAUCAAACGUCGGGCAUUCUU .....................((((((((......)))(((((...((((.(.((((((.......)))))).).))))...)))))))))) ( -20.90, z-score = -2.52, R) >droEre2.scaffold_4784 8576295 92 + 25762168 CCAAUUUUAGUUAAAAUUCUCAAGAAGGAUCAGACUCCUGCCUUUAGUGUAAAUAUCGCUCUGUAUGCGAUACUCAAACGACAGGCAAUCUU .....................((((((((......))))((((...((.....((((((.......)))))).....))...))))..)))) ( -19.40, z-score = -1.75, R) >consensus CCAAUUUUAGUUAAAAUUCUCAAGAAGGAUCAGACUCCUGCCUUCAGUUUGAAUAUCGCACAGUAUGCGAUACUCAAACGACAGGCAUUCUU .....................((((((((......))))((((...((((((.((((((.......)))))).))))))...))))..)))) (-19.90 = -19.90 + 0.00)

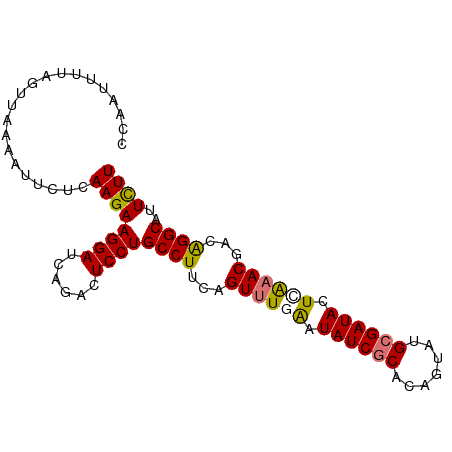
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:42 2011