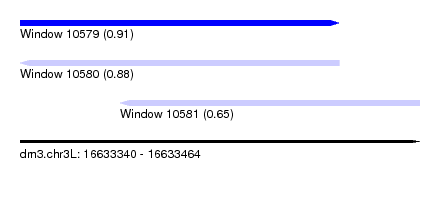
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,633,340 – 16,633,464 |
| Length | 124 |
| Max. P | 0.905585 |

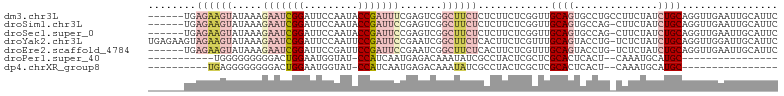
| Location | 16,633,340 – 16,633,439 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 62.95 |
| Shannon entropy | 0.65886 |
| G+C content | 0.46822 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -8.66 |
| Energy contribution | -8.10 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
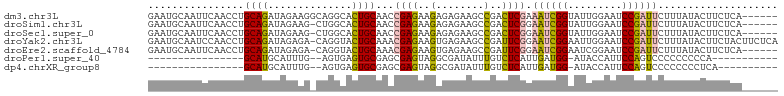
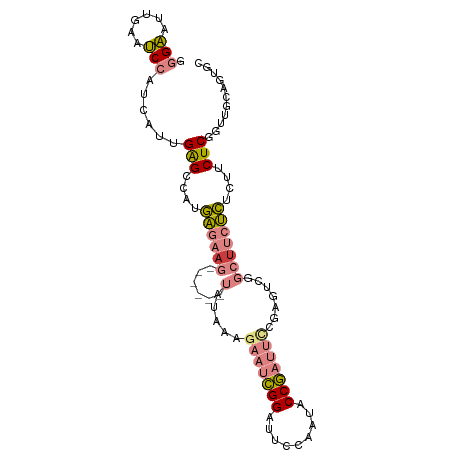
>dm3.chr3L 16633340 99 + 24543557 GAAUGCAAUUCAACCUGCAGAUAGAAGGCAGGCACUGCAACCGAGAAGAGAGAAGCCGACUCGAAAUCGGUAUUGGAAUCCGAUUCUUUAUACUUCUCA------ ...((((......(((((.........)))))...))))...(((((((((((((((((.......))))).((((...))))))))))...)))))).------ ( -27.00, z-score = -1.79, R) >droSim1.chr3L 15964060 98 + 22553184 GAAUGCAAUUCAACCUGCAGAUAGAAG-CUGGCACUGCAACCGAGAAGAGAGAAGCCGACUCGGAAUCGGUAUUGGAAUCCGAUUCUUUAUACUUCUCA------ ...((((......((.((........)-).))...))))...((((((.(((.......)))((((((((.........)))))))).....)))))).------ ( -25.90, z-score = -1.27, R) >droSec1.super_0 8711224 98 + 21120651 GAAUGCAAUUCAACCUGCAGAUAGAAG-CUGGCACUGCAACCGAGAAGAGAGAAGCCGACUCGGAAUCGGUAUUGGAAUCCGAUUCUUUAUACUUCUCA------ ...((((......((.((........)-).))...))))...((((((.(((.......)))((((((((.........)))))))).....)))))).------ ( -25.90, z-score = -1.27, R) >droYak2.chr3L 6760121 104 - 24197627 GAAUGCAAUCCAACCUGCAGAUAGAGA-CAGGUACUGCAAACGAGAAGUGAGAAGCCGAUUCGGAAUCGGAAUUGGAAUCCGAUUCUUUAUACUUCUACUUCUCA ...((((.....((((((.......).-)))))..))))...(((((((.(((((...((..(((((((((.......)))))))))..)).)))))))))))). ( -35.40, z-score = -4.19, R) >droEre2.scaffold_4784 8567156 98 - 25762168 GAAUGCAAUUCAACCUGCAGAUAGAGA-CAGGUACUGCAAACGAGAAGUGAGAAGCCGAUUCGGAAUCGGAAUCGGAAUCCGAUUCUUUAUACUUCUCA------ ...((((.....((((((.......).-)))))..))))...((((((((.((..(((...)))..))((((((((...))))))))...)))))))).------ ( -31.50, z-score = -3.28, R) >droPer1.super_40 700732 75 + 810552 ----------------GCAUGCAUUUG--AGUGAGUGCGAGCGAGUAGGCGAUAUUUGUCUCAUUGAUGG-AUACCAUUCCAGUCCCCCCCCCA----------- ----------------((.(((((((.--...))))))).))((((.((...((((..((.....))..)-)))))))))..............----------- ( -17.70, z-score = -0.91, R) >dp4.chrXR_group8 8492521 76 + 9212921 ----------------GCAUGCAUUUG--AGUGAGUGCGAGCGAGUAGGCGAUAUUUGUCUCAUUGAUGG-AUACCAUUCCAGUCCCCCCCCUCA---------- ----------------((.(((((((.--...))))))).))(((..((.(((....(((.....)))((-(......))).)))..))..))).---------- ( -20.00, z-score = -1.33, R) >consensus GAAUGCAAUUCAACCUGCAGAUAGAAG_CAGGCACUGCAACCGAGAAGAGAGAAGCCGACUCGGAAUCGGUAUUGGAAUCCGAUUCUUUAUACUUCUCA______ ................((((..............))))...((((..(........)..)))).((((((.........)))))).................... ( -8.66 = -8.10 + -0.57)
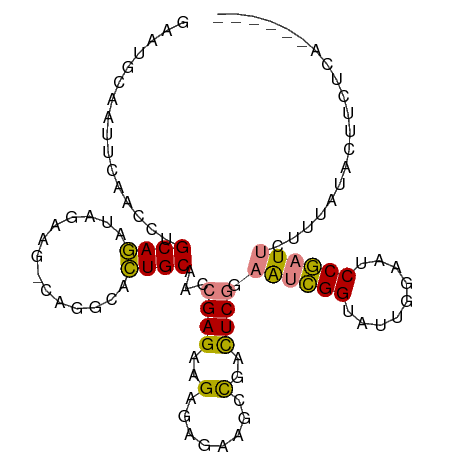
| Location | 16,633,340 – 16,633,439 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 62.95 |
| Shannon entropy | 0.65886 |
| G+C content | 0.46822 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -9.30 |
| Energy contribution | -11.94 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16633340 99 - 24543557 ------UGAGAAGUAUAAAGAAUCGGAUUCCAAUACCGAUUUCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCUGCCUUCUAUCUGCAGGUUGAAUUGCAUUC ------.(((((((.....(((((((.........))))))).......)))))))..........((((((((((((.........))))))...))))))... ( -29.20, z-score = -1.87, R) >droSim1.chr3L 15964060 98 - 22553184 ------UGAGAAGUAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCAG-CUUCUAUCUGCAGGUUGAAUUGCAUUC ------.(((((((.....((.(((((.((.......)).))))).)).)))))))..........((((((..(((-((((.....).)))))).))))))... ( -29.90, z-score = -2.03, R) >droSec1.super_0 8711224 98 - 21120651 ------UGAGAAGUAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCAG-CUUCUAUCUGCAGGUUGAAUUGCAUUC ------.(((((((.....((.(((((.((.......)).))))).)).)))))))..........((((((..(((-((((.....).)))))).))))))... ( -29.90, z-score = -2.03, R) >droYak2.chr3L 6760121 104 + 24197627 UGAGAAGUAGAAGUAUAAAGAAUCGGAUUCCAAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUACCUG-UCUCUAUCUGCAGGUUGGAUUGCAUUC .(((((((((((((.....((((((((.......)))))))).......)))))).)))))))...(((((((((((-(........))))))...))))))... ( -40.40, z-score = -5.39, R) >droEre2.scaffold_4784 8567156 98 + 25762168 ------UGAGAAGUAUAAAGAAUCGGAUUCCGAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUACCUG-UCUCUAUCUGCAGGUUGAAUUGCAUUC ------((((((((.....((((((((.......)))))))).......)))))))).........(((((((((((-(........))))))...))))))... ( -33.60, z-score = -4.00, R) >droPer1.super_40 700732 75 - 810552 -----------UGGGGGGGGGACUGGAAUGGUAU-CCAUCAAUGAGACAAAUAUCGCCUACUCGCUCGCACUCACU--CAAAUGCAUGC---------------- -----------.(((..((.((.((((......)-)))((.....))......)).))..)))((..(((......--....)))..))---------------- ( -13.50, z-score = 1.39, R) >dp4.chrXR_group8 8492521 76 - 9212921 ----------UGAGGGGGGGGACUGGAAUGGUAU-CCAUCAAUGAGACAAAUAUCGCCUACUCGCUCGCACUCACU--CAAAUGCAUGC---------------- ----------(((((.(((.((.(((..((((((-...((.....))...)))))).))).)).))).).))))..--...........---------------- ( -15.40, z-score = 0.73, R) >consensus ______UGAGAAGUAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGCCUG_CUUCUAUCUGCAGGUUGAAUUGCAUUC ........((((((.....(((((((.........))))))).......))))))..........((((((..............)))))).............. ( -9.30 = -11.94 + 2.64)
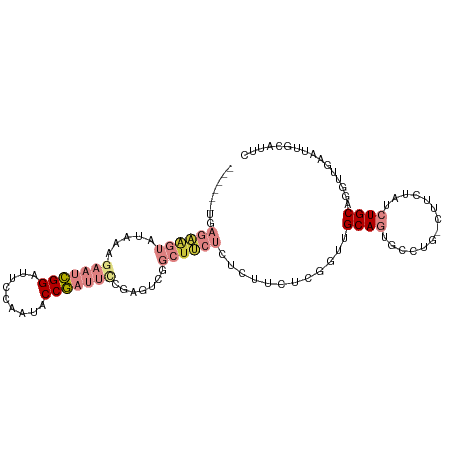
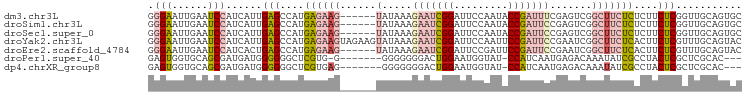
| Location | 16,633,371 – 16,633,464 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 68.75 |
| Shannon entropy | 0.55704 |
| G+C content | 0.48828 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -11.79 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.645764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16633371 93 - 24543557 GGGAAUUGAAUCCAUCAUUGAGCCAUGAGAAG------UAUAAAGAAUCGGAUUCCAAUACCGAUUUCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGC .(((......)))..(((((((((..((((((------.....((((.(.(((((.((.......)).))))).).))))..)))))))))).))))). ( -24.90, z-score = -0.71, R) >droSim1.chr3L 15964090 93 - 22553184 GGGAAUUGAAUCCAUCAUUGAGCCAUGAGAAG------UAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGC .(((......)))..(((((((((..((((((------(.....((.(((((.((.......)).))))).)).))))))).......)))).))))). ( -27.10, z-score = -1.35, R) >droSec1.super_0 8711254 93 - 21120651 GGGAAUUGAAUCCAUCAUUGAGCCAUGAGAAG------UAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGC .(((......)))..(((((((((..((((((------(.....((.(((((.((.......)).))))).)).))))))).......)))).))))). ( -27.10, z-score = -1.35, R) >droYak2.chr3L 6760151 99 + 24197627 GGGAAUUGAAUCCAUCAUUGAGCCAUGAGAAGUAGAAGUAUAAAGAAUCGGAUUCCAAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUAC .(((......)))........((.(((((((((((((((.....((((((((.......)))))))).......)))))).)))))))))..))..... ( -33.80, z-score = -3.91, R) >droEre2.scaffold_4784 8567186 93 + 25762168 GGGAAUUGAAUCCAUCACUGAGCCAUGAGAAG------UAUAAAGAAUCGGAUUCCGAUUCCGAUUCCGAAUCGGCUUCUCACUUCUCGUUUGCAGUAC .(((......)))...((((....((((((((------(.....(((((((...))))))).((..(((...)))..))..)))))))))...)))).. ( -28.60, z-score = -2.28, R) >droPer1.super_40 700748 87 - 810552 GAGUGGUGCAGCGAUGAUGGGGGGCUCGUG-G-------GGGGGGGACUGGAAUGGUAU-CCAUCAAUGAGACAAAUAUCGCCUACUCGCUCGCAC--- .....(((((((((.(.((((.(((((((.-.-------(..((..(((.....)))..-))..).))))).......)).)))))))))).))))--- ( -26.81, z-score = -0.36, R) >dp4.chrXR_group8 8492537 88 - 9212921 GAGUGGUGCAGCGAUGAUGGGGGGCUCGUGAG-------GGGGGGGACUGGAAUGGUAU-CCAUCAAUGAGACAAAUAUCGCCUACUCGCUCGCAC--- .....((((((((((((((((...(((....)-------))((....)).........)-))))))..((........))......))))).))))--- ( -27.80, z-score = -0.66, R) >consensus GGGAAUUGAAUCCAUCAUUGAGCCAUGAGAAG______UAUAAAGAAUCGGAUUCCAAUACCGAUUCCGAGUCGGCUUCUCUCUUCUCGGUUGCAGUGC .(((......)))......(((....((((((............(((((((.........)))))))((...)).))))))....)))........... (-11.79 = -11.90 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:36 2011