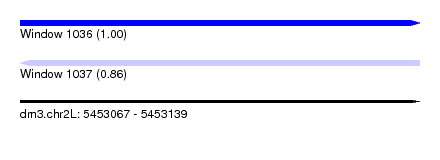
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,453,067 – 5,453,139 |
| Length | 72 |
| Max. P | 0.999542 |

| Location | 5,453,067 – 5,453,139 |
|---|---|
| Length | 72 |
| Sequences | 9 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 56.38 |
| Shannon entropy | 0.88107 |
| G+C content | 0.55933 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -7.68 |
| Energy contribution | -7.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
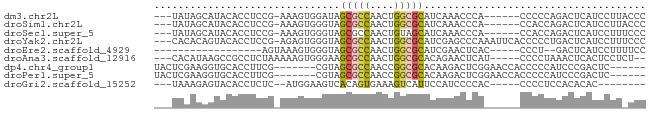
>dm3.chr2L 5453067 72 + 23011544 ---UAUAGCAUACACCUCCG-AAAGUGGAUAGCGCCAACUGGCGCAUCAAACCCA------CCCCCAGACUCAUCCUUACCC ---.........(((.....-...)))(((.(((((....)))))))).......------..................... ( -13.70, z-score = -2.25, R) >droSim1.chr2L 5255845 72 + 22036055 ---UAUAGCAUACACCUCCG-AAAGUGGGUAGCGCCAACUGGCGCAUCAAACCCA------CCACCAGACUCAUCCUUACCC ---.................-...((((((.(((((....))))).....)))))------).................... ( -18.10, z-score = -2.95, R) >droSec1.super_5 3527258 72 + 5866729 ---UAUAGCAUACACCUCCG-AAAGUGGGUAGCGCCAACUGUAGCAUCAAACCCA------CCACCAGACUCAUCCUUUCCC ---....((.((((((..(.-...)..)))((......)))))))..........------..................... ( -9.60, z-score = -0.19, R) >droYak2.chr2L 8580350 78 - 22324452 ---CACACAGUACACCUCCG-AGAGUGGGUAGCGCCAACUGGCGCAUCGAGCCCAAAUUCACCCCCUGACUCAUCCUUUCCC ---................(-((((.((((.(((((....)))))...(((......))))))).))..))).......... ( -19.80, z-score = -1.73, R) >droEre2.scaffold_4929 5537266 57 + 26641161 ------------------AGUAAAGUGGGUAGCGCCAACUGGCGCAUCGAACUCAC-----CCCU--GACUCAUCCUUUUCC ------------------(((..((.((((.(((((....)))))...(....)))-----))))--.)))........... ( -16.10, z-score = -1.87, R) >droAna3.scaffold_12916 6479557 72 - 16180835 ---CACAUAAGCCGCCUCUAAAAAGUGGGAAGCGCCAACUGGCGCACAGAACUCAU-----CCCCUAAACUCACUCCUCU-- ---....................((.((((.(((((....)))))...(....).)-----)))))..............-- ( -16.10, z-score = -1.99, R) >dp4.chr4_group1 4758334 69 + 5278887 UACUCGAAGGUGCACCUUCG-------CGUAGCGCCAACCGGCGCACAAGACUCGGAACCACCCCCAUCCCGACUC------ (((.((((((....))))))-------.)))(((((....)))))....((.((((.............)))).))------ ( -25.22, z-score = -3.99, R) >droPer1.super_5 6354019 69 - 6813705 UACUCGAAGGUGCACCUUCG-------CGUAGCGCCAACCGGCGCACAAGACUCGGAACCACCCCCAUCCCGACUC------ (((.((((((....))))))-------.)))(((((....)))))....((.((((.............)))).))------ ( -25.22, z-score = -3.99, R) >droGri2.scaffold_15252 5913488 64 - 17193109 ---UAAAGAGUACACCUCUC--AUGGAAGUCACAGUGAAAGUCAUUCCAUCCCCAC-----CCCCUCCACACAC-------- ---....(((.......)))--((((((...((.......))..))))))......-----.............-------- ( -6.90, z-score = -0.02, R) >consensus ___UAUAGAAUACACCUCCG_AAAGUGGGUAGCGCCAACUGGCGCAUCAAACCCA______CCCCCAGACUCAUCCUU_CCC ...............................(((((....)))))..................................... ( -7.68 = -7.88 + 0.20)

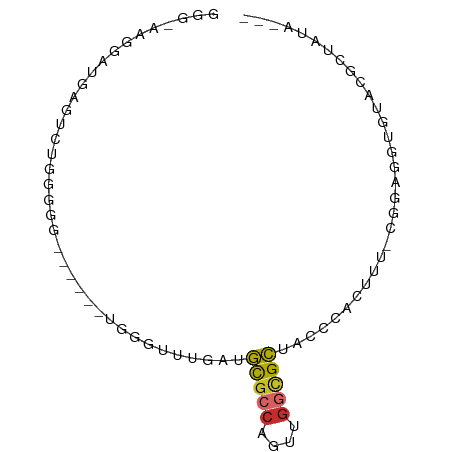
| Location | 5,453,067 – 5,453,139 |
|---|---|
| Length | 72 |
| Sequences | 9 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 56.38 |
| Shannon entropy | 0.88107 |
| G+C content | 0.55933 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -7.12 |
| Energy contribution | -7.19 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
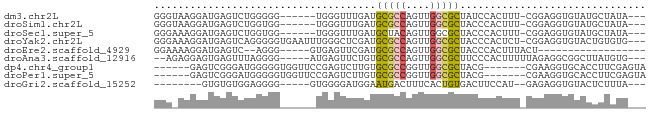
>dm3.chr2L 5453067 72 - 23011544 GGGUAAGGAUGAGUCUGGGGG------UGGGUUUGAUGCGCCAGUUGGCGCUAUCCACUUU-CGGAGGUGUAUGCUAUA--- .((((...(((..((((..((------(((((.....(((((....))))).)))))))..-))))..))).))))...--- ( -30.90, z-score = -3.99, R) >droSim1.chr2L 5255845 72 - 22036055 GGGUAAGGAUGAGUCUGGUGG------UGGGUUUGAUGCGCCAGUUGGCGCUACCCACUUU-CGGAGGUGUAUGCUAUA--- .((((...(((..(((((.((------(((((.....(((((....))))).))))))).)-))))..))).))))...--- ( -29.80, z-score = -3.09, R) >droSec1.super_5 3527258 72 - 5866729 GGGAAAGGAUGAGUCUGGUGG------UGGGUUUGAUGCUACAGUUGGCGCUACCCACUUU-CGGAGGUGUAUGCUAUA--- .....((.(((..(((((.((------(((((..(.(((((....)))))).))))))).)-))))....))).))...--- ( -22.50, z-score = -1.24, R) >droYak2.chr2L 8580350 78 + 22324452 GGGAAAGGAUGAGUCAGGGGGUGAAUUUGGGCUCGAUGCGCCAGUUGGCGCUACCCACUCU-CGGAGGUGUACUGUGUG--- (((......((((((..((......))..))))))..(((((....)))))..))).....-((.((.....)).))..--- ( -23.10, z-score = 0.54, R) >droEre2.scaffold_4929 5537266 57 - 26641161 GGAAAAGGAUGAGUC--AGGG-----GUGAGUUCGAUGCGCCAGUUGGCGCUACCCACUUUACU------------------ .....((...((((.--..((-----(((........(((((....))))))))))))))..))------------------ ( -15.90, z-score = -0.37, R) >droAna3.scaffold_12916 6479557 72 + 16180835 --AGAGGAGUGAGUUUAGGGG-----AUGAGUUCUGUGCGCCAGUUGGCGCUUCCCACUUUUUAGAGGCGGCUUAUGUG--- --......(((((((..((((-----(..........(((((....)))))))))).(((....)))..)))))))...--- ( -19.40, z-score = 0.27, R) >dp4.chr4_group1 4758334 69 - 5278887 ------GAGUCGGGAUGGGGGUGGUUCCGAGUCUUGUGCGCCGGUUGGCGCUACG-------CGAAGGUGCACCUUCGAGUA ------((.(((((((.......))))))).))....(((((....)))))(((.-------((((((....)))))).))) ( -30.50, z-score = -2.04, R) >droPer1.super_5 6354019 69 + 6813705 ------GAGUCGGGAUGGGGGUGGUUCCGAGUCUUGUGCGCCGGUUGGCGCUACG-------CGAAGGUGCACCUUCGAGUA ------((.(((((((.......))))))).))....(((((....)))))(((.-------((((((....)))))).))) ( -30.50, z-score = -2.04, R) >droGri2.scaffold_15252 5913488 64 + 17193109 --------GUGUGUGGAGGGG-----GUGGGGAUGGAAUGACUUUCACUGUGACUUCCAU--GAGAGGUGUACUCUUUA--- --------....(((((((..-----(..((((.(......).))).)..)..)))))))--(((((.....)))))..--- ( -18.60, z-score = -1.17, R) >consensus GGG_AAGGAUGAGUCUGGGGG______UGGGUUUGAUGCGCCAGUUGGCGCUACCCACUUU_CGGAGGUGUACGCUAUA___ .....................................(((((....)))))............................... ( -7.12 = -7.19 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:05 2011