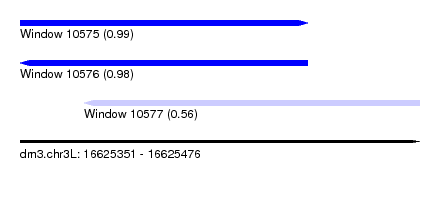
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,625,351 – 16,625,476 |
| Length | 125 |
| Max. P | 0.993658 |

| Location | 16,625,351 – 16,625,441 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.37200 |
| G+C content | 0.52165 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -19.78 |
| Energy contribution | -22.25 |
| Covariance contribution | 2.47 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
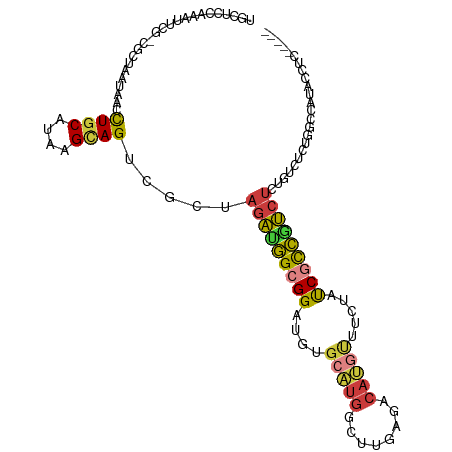
>dm3.chr3L 16625351 90 + 24543557 UGUUGGGUGAGAAAGAGUCGGAGGUAUGGCCAGAGAAAGAGACGGCGAUAGAAACAUGUCUCAAGCCAUGCACAUCCGCCAUCUAGCGAC .((((((((.((.....))(((.(((((((..((((.........(....).......))))..)))))))...)))..))))))))... ( -28.39, z-score = -2.32, R) >droSim1.chr3L 15956521 90 + 22553184 UGUUGGGUGGGAAAGAGGCGGAGGUAUGGCCAGAGACAGAGACGGCGAUAGAAACAUGUCUCAAGACAUGCACAUCCGCCAUCUAGCGAC .(((((((........((((((.(((((....((((((.......(....).....))))))....)))))...)))))))))))))... ( -30.30, z-score = -2.79, R) >droSec1.super_0 8703259 90 + 21120651 UGUUGGGUGGGAAAGAGGCGGAGGUAUGGCCAGAGACAGAGACGGCGAUAGAAACAUGUCUCAAGCCAUGCACAUCCGCCAUCUAGAGAC .............(((((((((.(((((((..((((((.......(....).....))))))..)))))))...)))))).)))...... ( -34.70, z-score = -4.12, R) >droYak2.chr3L 6752027 90 - 24197627 UGUUGGGAGGGAAAGAGGCGGAGGUAUGGCUGGAGACAGAGAUGGCGAUAGAAACAUGUCUCAAGCCAUGCACAUCCGCCAUCUAGCGAC ..........(..(((((((((.((((((((.((((((.......(....).....)))))).))))))))...)))))).)))..)... ( -37.40, z-score = -4.75, R) >droEre2.scaffold_4784 8559116 90 - 25762168 UGUUGGGAGAGAAAGAGGCGGAUAUAUGGAUGGAGACAGAGAUGGCGAUAGAAACAUGUCUCAAGCCAUGCACAUCCGCCAUCUAGCGAC .((((..(((......(((((((.(((((...((((((.......(....).....))))))...)))))...))))))).)))..)))) ( -29.50, z-score = -3.30, R) >droAna3.scaffold_13337 9323286 78 - 23293914 ------------UGCUUAGAGAAGAAAAAAGAAAGAGAGAGAAGGGGAGAAGACACUGUCUCAAGUCAUGCACAUCCGCCAUCUAGCGGC ------------.(((.(((..........((..((((.((...(........).)).))))...))..((......))..))))))... ( -12.50, z-score = 1.06, R) >consensus UGUUGGGUGGGAAAGAGGCGGAGGUAUGGCCAGAGACAGAGACGGCGAUAGAAACAUGUCUCAAGCCAUGCACAUCCGCCAUCUAGCGAC .............(((((((((.(((((((..((((((..................))))))..)))))))...)))))).)))...... (-19.78 = -22.25 + 2.47)



| Location | 16,625,351 – 16,625,441 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.37200 |
| G+C content | 0.52165 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -18.72 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.977034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16625351 90 - 24543557 GUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCGUCUCUUUCUCUGGCCAUACCUCCGACUCUUUCUCACCCAACA ((((..(((((((((....(((((........))))).....)))))))))........((.....))..))))................ ( -23.00, z-score = -1.14, R) >droSim1.chr3L 15956521 90 - 22553184 GUCGCUAGAUGGCGGAUGUGCAUGUCUUGAGACAUGUUUCUAUCGCCGUCUCUGUCUCUGGCCAUACCUCCGCCUCUUUCCCACCCAACA ...(..(((.((((((.(((((((((....))))))).......((((..........))))...)).)))))))))..).......... ( -25.80, z-score = -2.32, R) >droSec1.super_0 8703259 90 - 21120651 GUCUCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCGUCUCUGUCUCUGGCCAUACCUCCGCCUCUUUCCCACCCAACA ......(((.((((((.((..((((((.((((((.(...(.......)...))))))).)))))))).)))))))))............. ( -30.00, z-score = -3.54, R) >droYak2.chr3L 6752027 90 + 24197627 GUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCAUCUCUGUCUCCAGCCAUACCUCCGCCUCUUUCCCUCCCAACA ...(..(((.((((((.((..((((((.((((((..................)))))).)))))))).)))))))))..).......... ( -30.17, z-score = -3.90, R) >droEre2.scaffold_4784 8559116 90 + 25762168 GUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCAUCUCUGUCUCCAUCCAUAUAUCCGCCUCUUUCUCUCCCAACA ...(..(((.(((((((((..((((...((((((..................))))))...))))))))))))))))..).......... ( -26.57, z-score = -3.08, R) >droAna3.scaffold_13337 9323286 78 + 23293914 GCCGCUAGAUGGCGGAUGUGCAUGACUUGAGACAGUGUCUUCUCCCCUUCUCUCUCUUUCUUUUUUCUUCUCUAAGCA------------ .(((((....)))))........((...((((.((..............)).))))..))..................------------ ( -10.64, z-score = 1.75, R) >consensus GUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCGUCUCUGUCUCUGGCCAUACCUCCGCCUCUUUCCCACCCAACA ......(((.((((((.((..((((((.((((((..................)))))).)))))))).)))))))))............. (-18.72 = -20.00 + 1.28)


| Location | 16,625,371 – 16,625,476 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.36 |
| Shannon entropy | 0.47987 |
| G+C content | 0.49794 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.23 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16625371 105 - 24543557 UGCUCCAAAUUCGCCGCUAAUAACCUGCAUAAGCAGUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCGUCUCUUUCUCUGGCCAUACCUC----- .((.....(((((((((((.....((((....))))....))).))))))))..))((((((.(((((..............))))).......)))))).....----- ( -30.54, z-score = -1.50, R) >droSim1.chr3L 15956541 105 - 22553184 UGCUCCAAAUUCGCCGCUAAUAACCUGCAUAAGCAGUCGCUAGAUGGCGGAUGUGCAUGUCUUGAGACAUGUUUCUAUCGCCGUCUCUGUCUCUGGCCAUACCUC----- ............(((.........((((....))))..((.(((((((((....(((((((....))))))).....)))))))))..))....)))........----- ( -32.40, z-score = -2.40, R) >droSec1.super_0 8703279 105 - 21120651 UGCUCCAAAUUCGCCGCUAAUAACCUGCAUAAGCAGUCUCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCGUCUCUGUCUCUGGCCAUACCUC----- .((.....(((((((((((.....((((....))))....))).))))))))..))((((((.((((((.(...(.......)...))))))).)))))).....----- ( -34.70, z-score = -3.01, R) >droYak2.chr3L 6752047 104 + 24197627 UGCUCCAAAUUCA-CGCUAAUAACCUGCAUAAGCAGUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCAUCUCUGUCUCCAGCCAUACCUC----- .((.(.....((.-(((((.....((((....))))((....))))))))).).))((((((.((((((..................)))))).)))))).....----- ( -27.67, z-score = -1.07, R) >droEre2.scaffold_4784 8559136 104 + 25762168 UGCUCCAAAUUCA-CGCUAAUAACCUGCAUAAGCAGUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCAUCUCUGUCUCCAUCCAUAUAUC----- .............-..........((((....))))..((.(((((((((....(((((........))))).....)))))))))..))...............----- ( -24.00, z-score = -0.33, R) >droAna3.scaffold_13337 9323305 93 + 23293914 UGCUCCAAAUUCC-CGCUAAUAGCCUCCAUAAGCAGCCGCUAGAUGGCGGAUGUGCAUGACUUGAGACAGUGUCUUCUCCCCUUCUCUCUCUUU---------------- .............-.((.....))...(((..(((.(((((....))))).)))..)))....((((.((..............)).))))...---------------- ( -16.24, z-score = 1.22, R) >droWil1.scaffold_180727 2506530 107 + 2741493 UGCUCCAAUUCC--UACUAAUAACUUGCAUAAGUAGCUAUAAUGGCGCGGAGAUGUGACGCUGAAGAUGC-CUCCUCCCAUGCCUCAUGCCGCAUGCCGCAUGCCCGAAU .((.(((((..(--((((.((.......)).)))))..))..))).))((((....((.((.......))-.))))))(((((..((((...))))..)))))....... ( -22.00, z-score = 0.84, R) >consensus UGCUCCAAAUUCG_CGCUAAUAACCUGCAUAAGCAGUCGCUAGAUGGCGGAUGUGCAUGGCUUGAGACAUGUUUCUAUCGCCGUCUCUGUCUCUGGCCAUACCUC_____ ........................((((....)))).....(((((((((....(((((........))))).....)))))))))........................ (-17.10 = -17.23 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:33 2011