| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,564,313 – 16,564,453 |
| Length | 140 |
| Max. P | 0.962743 |

| Location | 16,564,313 – 16,564,410 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 90.89 |
| Shannon entropy | 0.17280 |
| G+C content | 0.41020 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
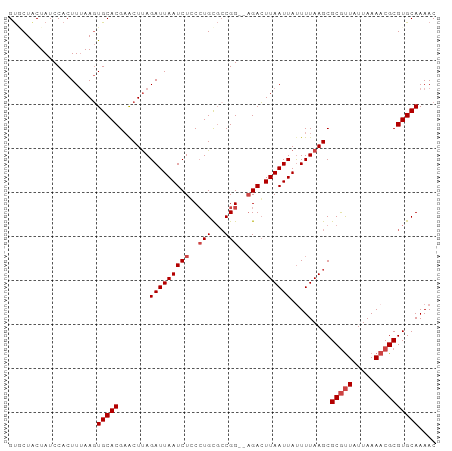
>dm3.chr3L 16564313 97 + 24543557 GUGCUACUAUCCACUUUAAGUGCACGAACUUAGAUUAAUCUGCCUGCGACGGAUAGACUUAAUUAUUUUAAGCGCGUUAUUAAAACGCGUGCAAAAC .(((..((((((..(((((((......)))))))......((....))..))))))...............(((((((.....)))))))))).... ( -24.20, z-score = -2.33, R) >droSim1.chr3L 15909558 97 + 22553184 GUCCUACUAUCCACUUUAAGUGCACGAACUUAGAUUAAUCUGCCUGCGCCGGAUAGACUUAAUUAUUUUAAGCGCGUUAUUAAAACGCGUGCAAAAC ......((((((..(((((((......))))))).......((....)).)))))).........((((..(((((((.....)))))))..)))). ( -23.30, z-score = -2.36, R) >droSec1.super_0 8656508 97 + 21120651 GUCCUACUAUCCACUUUAAGUGCACGAACUUAGAUUAAUCUGCCUGCGCCGGAUAGACUUAAUUAUUUUAAGCGCGUUAUUAAAACGCGUGCAAAAC ......((((((..(((((((......))))))).......((....)).)))))).........((((..(((((((.....)))))))..)))). ( -23.30, z-score = -2.36, R) >droYak2.chr3L 6703935 95 - 24197627 GUGCUACUAUCCACUUAAAGUGCACGAACUCAGAUUAAUCUCCCUGCGCCGG--AGACUUAAUUAUUUUAAGCGCGUUAUUAAAACGCGUGCAAAAC ((((.(((..........))))))).......(((((((((((.......))--))).)))))).((((..(((((((.....)))))))..)))). ( -24.50, z-score = -2.82, R) >droEre2.scaffold_4784 8510517 95 - 25762168 GUGCUGCUAUCCACUUUAAGUGCACGAACUUAGAUUAAUCUCCCUGCGCCGG--AGACUUAAUUACUUUAAGCGCGUUGUUAAAACGCGUGCAAAAC (..(.(((........(((((......)))))(((((((((((.......))--))).))))))......)))(((((.....))))))..)..... ( -24.80, z-score = -1.95, R) >droAna3.scaffold_13337 9282021 92 - 23293914 GCACUACUAUCCACUUUAAUUGCACGUGCUUAGAUUAAUCUCCGUGU-CCGG---GAGUUAAUUAUUUUAAGCGCACUUUUAAA-CGCGUGCAAAAC ...................((((((((((((((((((((..(((...-.)))---..)))))))....))))))).(.......-.).))))))... ( -23.50, z-score = -2.77, R) >consensus GUGCUACUAUCCACUUUAAGUGCACGAACUUAGAUUAAUCUCCCUGCGCCGG__AGACUUAAUUAUUUUAAGCGCGUUAUUAAAACGCGUGCAAAAC ....................(((((.......(((((((((..(((...)))..))).)))))).........(((((.....)))))))))).... (-15.08 = -15.75 + 0.67)


| Location | 16,564,349 – 16,564,445 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 65.36 |
| Shannon entropy | 0.62403 |
| G+C content | 0.46319 |
| Mean single sequence MFE | -24.16 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.71 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
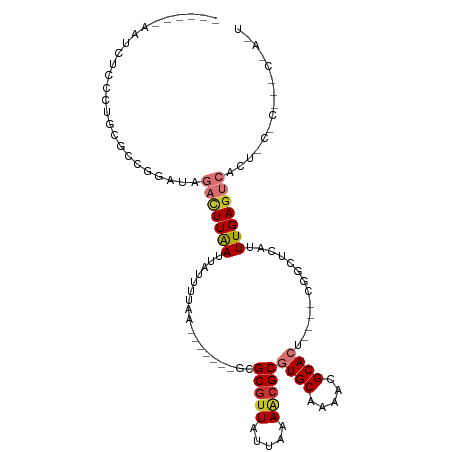
>dm3.chr3L 16564349 96 + 24543557 ------AAUCUGCCUGCGACGGAUAGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CAACUCAUUUGAGUCGCUCCACACUCCACU ------..............(((..(((((((.........-------..(((((.....)))))((((.....)))).----........)))))))..))).......... ( -22.50, z-score = -2.20, R) >droSim1.chr3L 15909594 96 + 22553184 ------AAUCUGCCUGCGCCGGAUAGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGUCGCUCCACACUCCACU ------..((((((......)).))))(((((.....))))-------).(((((.....)))))((((.....)))).----((((((....)))))).............. ( -23.90, z-score = -1.48, R) >droSec1.super_0 8656544 96 + 21120651 ------AAUCUGCCUGCGCCGGAUAGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGUCGCUCCACACUCCACU ------..((((((......)).))))(((((.....))))-------).(((((.....)))))((((.....)))).----((((((....)))))).............. ( -23.90, z-score = -1.48, R) >droYak2.chr3L 6703971 82 - 24197627 ------AAUCUCCCUGCGCCGG--AGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGUCACU------------ ------..(((((.......))--)))(((((.....))))-------).(((((.....)))))((((.....)))).----.(((((....)))))...------------ ( -24.70, z-score = -2.84, R) >droEre2.scaffold_4784 8510553 82 - 25762168 ------AAUCUCCCUGCGCCGG--AGACUUAAUUACUUUAA-------GCGCGUUGUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGUCACU------------ ------..(((((.......))--)))(((((.....))))-------).(((((.....)))))((((.....)))).----.(((((....)))))...------------ ( -25.60, z-score = -2.72, R) >droAna3.scaffold_13337 9282057 81 - 23293914 ------AAUCUCCGUGU-CCGG---GAGUUAAUUAUUUUAA-------GCGCACUUUUAAA-CGCGUGCAAAACGCACUGAGCCAGCCCAUUUGAAUCG-------------- ------...........-..((---(.(((...........-------(((..........-)))((((.....))))..)))...)))..........-------------- ( -13.50, z-score = 0.89, R) >dp4.chrXR_group8 2266770 113 + 9212921 AAUUUUGUCCGCUGUCCAGCUAAUUAUUUUGAUUGUUUGCAUGGAGGCGUGCGUUUUUUAAGCGCGUGCAAAACGCAGCACUAGCCCCGUUUUGAGUCAGAGCCCGUCCGAAU ...((((..((((....)))......((((((((......((((.((((((((((.....)))((((.....)))).))))..)))))))....))))))))...)..)))). ( -29.40, z-score = -0.32, R) >droPer1.super_24 767185 113 - 1556852 AAUUUUGUCCGCUGUCCAGCUAAUUAUUUUGAUUGUUUGCAUGGAGGCGUGCGUUUUUUAAGCGCGUGCAAAACGCAGCACUAGCCCCAUUUUGAGUCAGAGCCCGUCCGAAU ...((((..((((....)))......((((((((......((((.((((((((((.....)))((((.....)))).))))..)))))))....))))))))...)..)))). ( -29.80, z-score = -0.67, R) >consensus ______AAUCUCCCUGCGCCGGAUAGACUUAAUUAUUUUAA_______GCGCGUUAUUAAAACGCGUGCAAAACGCACU____CGGCUCAUUUGAGUCACU_C_C___C_A_U .........................(((((((..................(((((.....)))))((((.....)))).............)))))))............... (-13.09 = -13.71 + 0.63)


| Location | 16,564,357 – 16,564,453 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.28 |
| Shannon entropy | 0.63780 |
| G+C content | 0.46676 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -10.43 |
| Energy contribution | -11.48 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.831130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16564357 96 + 24543557 -------UGCGACGGAUAGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CAACUCAUUUGAGUCGCUCCACACUCCACUCAGGUGAU -------......(((..(((((((.........-------..(((((.....)))))((((.....)))).----........)))))))..))).((((.......)))).. ( -24.80, z-score = -2.24, R) >droSim1.chr3L 15909602 96 + 22553184 -------UGCGCCGGAUAGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGUCGCUCCACACUCCACUCAGGUGAU -------...(((((...(.(((((.....))))-------))(((((.....)))))((((.....)))))----))))((((((((((.............)))))))))). ( -29.92, z-score = -2.84, R) >droSec1.super_0 8656552 96 + 21120651 -------UGCGCCGGAUAGACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGUCGCUCCACACUCCACUCAGGUGAU -------...(((((...(.(((((.....))))-------))(((((.....)))))((((.....)))))----))))((((((((((.............)))))))))). ( -29.92, z-score = -2.84, R) >droYak2.chr3L 6703979 82 - 24197627 -------UGCGCCGGA--GACUUAAUUAUUUUAA-------GCGCGUUAUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGU------------CACUCAGGUGAU -------...(((((.--(.(((((.....))))-------))(((((.....)))))((((.....)))))----))))(((((((((.------------..))))))))). ( -28.70, z-score = -3.33, R) >droEre2.scaffold_4784 8510561 82 - 25762168 -------UGCGCCGGA--GACUUAAUUACUUUAA-------GCGCGUUGUUAAAACGCGUGCAAAACGCACU----CGGCUCAUUUGAGU------------CACUCAGAUGAU -------...(((((.--(.(((((.....))))-------))(((((.....)))))((((.....)))))----))))(((((((((.------------..))))))))). ( -30.10, z-score = -3.61, R) >droAna3.scaffold_13337 9282065 81 - 23293914 ---------UGUCCGG--GAGUUAAUUAUUUUAA-------GCGCACU-UUUAAACGCGUGCAAAACGCACUGAGCCAGCCCAUUUGAAU--------------CGUGAUUGGU ---------.....((--(.(((...........-------(((....-......)))((((.....))))..)))...)))........--------------.......... ( -13.50, z-score = 1.68, R) >dp4.chrXR_group8 2266777 114 + 9212921 UCCGCUGUCCAGCUAAUUAUUUUGAUUGUUUGCAUGGAGGCGUGCGUUUUUUAAGCGCGUGCAAAACGCAGCACUAGCCCCGUUUUGAGUCAGAGCCCGUCCGAAUGAGGUGUG .(((((....)))............(..((((.((((.((((((((((.....)))((((.....)))).))))..)))..((((((...)))))))))).))))..))).... ( -30.40, z-score = 0.29, R) >droPer1.super_24 767192 114 - 1556852 UCCGCUGUCCAGCUAAUUAUUUUGAUUGUUUGCAUGGAGGCGUGCGUUUUUUAAGCGCGUGCAAAACGCAGCACUAGCCCCAUUUUGAGUCAGAGCCCGUCCGAAUAAGGUGUG .(((((....)))...((((((.(((.((((.(((((.((((((((((.....)))((((.....)))).))))..))))))...)).....))))..))).)))))))).... ( -28.90, z-score = 0.33, R) >consensus _______UGCGCCGGAU_GACUUAAUUAUUUUAA_______GCGCGUUAUUAAAACGCGUGCAAAACGCACU____CGGCUCAUUUGAGUC____C_C___CCACUCAGGUGAU ...........................................(((((.....)))))((((.....)))).........((((((((.................)))))))). (-10.43 = -11.48 + 1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:30 2011