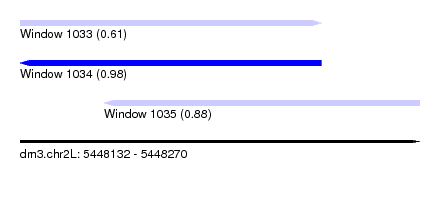
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,448,132 – 5,448,270 |
| Length | 138 |
| Max. P | 0.980103 |

| Location | 5,448,132 – 5,448,236 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Shannon entropy | 0.46608 |
| G+C content | 0.41741 |
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -12.77 |
| Energy contribution | -13.15 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
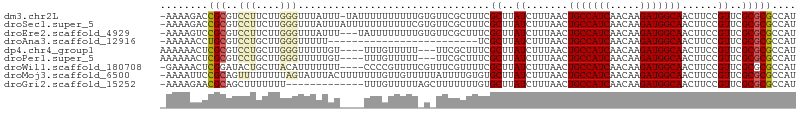
>dm3.chr2L 5448132 104 + 23011544 -AAAAGACCGCGUCCUUCUUGGGUUUAUUU-UAUUUUUUUUUUGUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -........(((((((....))).......-............(((..(((....))...........(((((((.....)))))))......)..)))))))... ( -20.90, z-score = -0.94, R) >droSec1.super_5 3522428 105 + 5866729 -AAAAGACCGCGUCCUUCUUGGGUUUAUUUAUUUUUUUUUUUCGUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -........(((((((....)))....................(((..(((....))...........(((((((.....)))))))......)..)))))))... ( -21.20, z-score = -1.00, R) >droEre2.scaffold_4929 5532316 102 + 26641161 -AAAAGUCCGCGUCCUUCUUGGGUUUAUUU---UAUUUUUUUUGUGUUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -........(((((((....))).......---..........(((..(((....))...........(((((((.....)))))))......)..)))))))... ( -20.90, z-score = -1.18, R) >droAna3.scaffold_12916 6474548 80 - 16180835 -AAAAACCUGCGUCCUGCUUGGGUUUUU-------------------------UCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -((((((((((.....))..))))))))-------------------------.(((...........(((((((.....)))))))......(....)))).... ( -18.90, z-score = -1.28, R) >dp4.chr4_group1 4751693 99 + 5278887 AAAAAACUCGCGUCCUGCUUGGGUUUUUGU----UUUGUUUUU---UUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU ........(((((...((..(((((.....----.........---...((....))............((((((.....)))))))))))..))..))))).... ( -20.50, z-score = -0.55, R) >droPer1.super_5 6347319 99 - 6813705 AAAAAACUCGCGUCCUGCUUGGGUUUUUGU----UUUGUUUUU---UUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU ........(((((...((..(((((.....----.........---...((....))............((((((.....)))))))))))..))..))))).... ( -20.50, z-score = -0.55, R) >droWil1.scaffold_180708 598339 101 + 12563649 -GAAAACUCGGAUACUGCUUACAUUUUUUU----CCCCGUUUUCGUUUCGUUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -((((((..(((.................)----))..))))))..........(((...........(((((((.....)))))))......(....)))).... ( -19.73, z-score = -1.64, R) >droMoj3.scaffold_6500 23376658 105 + 32352404 -AAAAUUCCGCAGUUUUUUUUAGUAUUUACUUUUUUUGUUGUUUUUAUUUUGUGUGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -.(((..(.((((........(((....)))....)))).)..))).....((((((..((.......(((((((.....)))))))......))..))))))... ( -18.72, z-score = -1.41, R) >droGri2.scaffold_15252 5905610 92 - 17193109 -AAAAGAACGCAGCUUUUUUU-------------UUUGUUUUUAGCUUUUUUUGUGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU -........((((((......-------------.........))))......((((..((.......(((((((.....)))))))......))..))))))... ( -18.08, z-score = -0.65, R) >consensus _AAAAACCCGCGUCCUUCUUGGGUUUUUUU____UUUGUUUUU_U_UUCGCUUUCGCUUAUCUUUAACUGCCAUCAACAAGAUGGCAACUUCCGUUCGCGCGCCAU ........(((..(((....)))................................((..((.......(((((((.....)))))))......))..))))).... (-12.77 = -13.15 + 0.39)
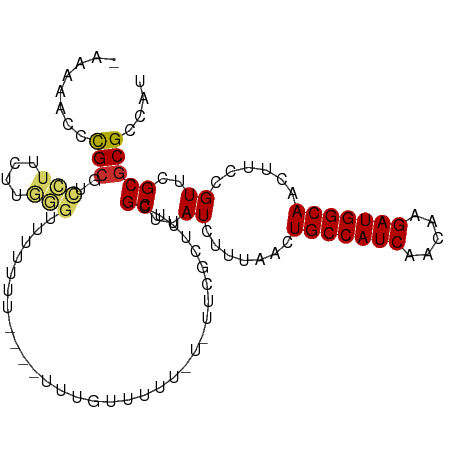
| Location | 5,448,132 – 5,448,236 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.35 |
| Shannon entropy | 0.46608 |
| G+C content | 0.41741 |
| Mean single sequence MFE | -25.41 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5448132 104 - 23011544 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAAAAAAAUA-AAAUAAACCCAAGAAGGACGCGGUCUUUU- ..(((.(((((((....)))((((((.....))))))............((....))..................-........((.....)).)))))))....- ( -28.30, z-score = -2.51, R) >droSec1.super_5 3522428 105 - 5866729 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACGAAAAAAAAAAAUAAAUAAACCCAAGAAGGACGCGGUCUUUU- ..(((.(((((((....)))((((((.....))))))............((....))...........................((.....)).)))))))....- ( -28.30, z-score = -2.30, R) >droEre2.scaffold_4929 5532316 102 - 26641161 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAAAAAUA---AAAUAAACCCAAGAAGGACGCGGACUUUU- ......(((((((....)))((((((.....))))))............((....))................---........((.....)).)))).......- ( -25.10, z-score = -1.77, R) >droAna3.scaffold_12916 6474548 80 + 16180835 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGA-------------------------AAAAACCCAAGCAGGACGCAGGUUUUU- ......(((.(((....)))((((((.....))))))............))).-------------------------(((((((...((.....)).)))))))- ( -23.60, z-score = -1.38, R) >dp4.chr4_group1 4751693 99 - 5278887 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAA---AAAAACAAA----ACAAAAACCCAAGCAGGACGCGAGUUUUUU ..(((.(((((((....)))((((((.....))))))............((....))...---.........----........((.....)).)))).))).... ( -26.50, z-score = -1.57, R) >droPer1.super_5 6347319 99 + 6813705 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAA---AAAAACAAA----ACAAAAACCCAAGCAGGACGCGAGUUUUUU ..(((.(((((((....)))((((((.....))))))............((....))...---.........----........((.....)).)))).))).... ( -26.50, z-score = -1.57, R) >droWil1.scaffold_180708 598339 101 - 12563649 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAACGAAACGAAAACGGGG----AAAAAAAUGUAAGCAGUAUCCGAGUUUUC- .((...(((.(((....)))((((((.....))))))............)))....))....((((((..((----(.(....((....)).).)))..))))))- ( -26.10, z-score = -1.54, R) >droMoj3.scaffold_6500 23376658 105 - 32352404 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCACACAAAAUAAAAACAACAAAAAAAGUAAAUACUAAAAAAAACUGCGGAAUUUU- ....(((((.(((....)))((((((.....))))))............))........................(((....)))..........))).......- ( -20.50, z-score = -1.51, R) >droGri2.scaffold_15252 5905610 92 + 17193109 AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCACAAAAAAAGCUAAAAACAAA-------------AAAAAAAGCUGCGUUCUUUU- ..(((((((.(((....)))((((((.....))))))............))........((((.........-------------......))))))))).....- ( -23.76, z-score = -1.62, R) >consensus AUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAA_A_AAAAACAAA____AAAAAAACCCAAGAAGGACGCGGGCUUUU_ ......(((((((....)))((((((.....)))))).........................................................))))........ (-18.52 = -18.52 + -0.00)

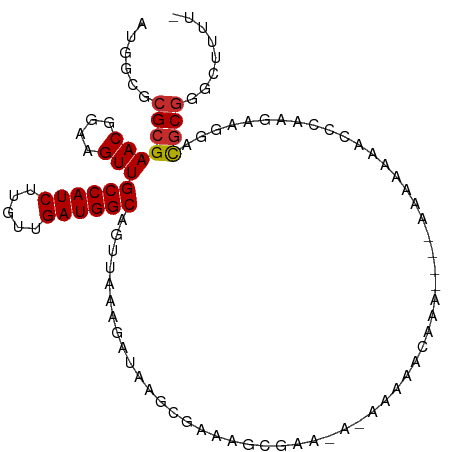
| Location | 5,448,161 – 5,448,270 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.61 |
| Shannon entropy | 0.41795 |
| G+C content | 0.42519 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.882170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
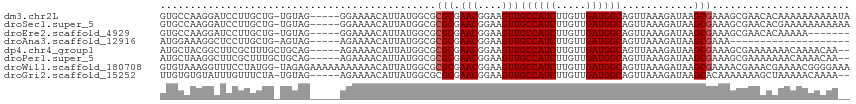
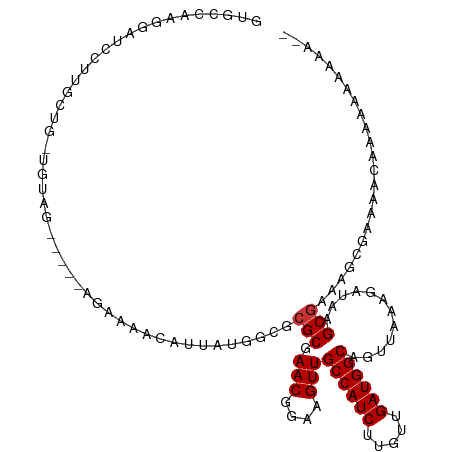
>dm3.chr2L 5448161 109 - 23011544 GUGCCAAGGAUCCUUGCUG-UGUAG-----GGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAAAAAAAUA ((((((.(..(((((((..-.))))-----)))...)....))))))...(((....)))((((((.....))))))............((....)).................. ( -33.30, z-score = -2.88, R) >droSec1.super_5 3522458 109 - 5866729 GUGCCAAGGAUCCUUGCUG-UGUAG-----GGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACGAAAAAAAAAAA ((((((.(..(((((((..-.))))-----)))...)....)))))).(((((....)))((((((.....))))))............((....)).....))........... ( -33.60, z-score = -2.93, R) >droEre2.scaffold_4929 5532350 102 - 26641161 GUGCCAAGGAUCCUUGCUG-UGUAG-----GGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAACACAAAAA------- ((((((.(..(((((((..-.))))-----)))...)....))))))...(((....)))((((((.....))))))............((....))...........------- ( -33.30, z-score = -2.72, R) >droAna3.scaffold_12916 6474573 89 + 16180835 AUGGAAAGGCUCCUUGCUG-AGUAG-----AGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAA-------------------- ........((((......)-)))..-----................(((.(((....)))((((((.....))))))............)))...-------------------- ( -21.10, z-score = -0.13, R) >dp4.chr4_group1 4751719 108 - 5278887 AUGCUACGGCUUCGCUUUGCUGCAG-----AGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAAAAAAACAAAACAA-- ..((....))(((((((((((..((-----.(....).))..))).(((.(((....)))((((((.....))))))............))))))))))).............-- ( -31.80, z-score = -1.66, R) >droPer1.super_5 6347345 108 + 6813705 AUGCUAAGGCUUCGCUUUGCUGCAG-----AGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAAAAAAACAAAACAA-- .((((...((((..(((((((((..-----....((((..((((((.....((....))))))))..))))....)))).)))))..))))...))))...............-- ( -30.90, z-score = -1.50, R) >droWil1.scaffold_180708 598365 114 - 12563649 GUGUAAAGGUUUCCUAUGG-UAGAGAAAAAAAAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAACGAAACGAAAACGGGGAAA ........(((((((((((-(................)))))))..(((.(((....)))((((((.....))))))............))).....)))))............. ( -23.79, z-score = -0.77, R) >droGri2.scaffold_15252 5905629 107 + 17193109 UUGUGUGUAUUUGUUUCUA-UGUAG-----AGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCACAAAAAAAGCUAAAAACAAAA-- ((((((((...(((((((.-.....-----)).))))).....))))))))((....))(((((((.....)))))))..........(((.........)))..........-- ( -26.50, z-score = -1.89, R) >consensus GUGCCAAGGAUCCUUGCUG_UGUAG_____AGAAAACAUUAUGGCGCGCGAACGGAAGUUGCCAUCUUGUUGAUGGCAGUUAAAGAUAAGCGAAAGCGAAAACAAAAAAAAAA__ ..............................................(((.(((....)))((((((.....))))))............)))....................... (-17.45 = -17.57 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:04 2011