
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,449,262 – 16,449,394 |
| Length | 132 |
| Max. P | 0.625368 |

| Location | 16,449,262 – 16,449,394 |
|---|---|
| Length | 132 |
| Sequences | 4 |
| Columns | 144 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.47076 |
| G+C content | 0.57665 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16449262 132 + 24543557 GACGCCUCGGGUUGUCCGCCUGUCGCUCAUUAGCGGCGCCCCCAAAGUCGCCGCUGAUGUGGCUGCUCCUGCUGCAGUUGCUCCACCUCCUCCUCC------------AGUCCAUCAAGUAUUUGUUAUUAAAUAUACGUGUGG ((.((..(((((.....)))))..))))((((((((((.(......).))))))))))(..(((((.......)))))..)...............------------...(((.((.(((((((....)))))))...))))) ( -42.20, z-score = -2.69, R) >droSim1.chr3L 15790715 138 + 22553184 GACGCCUCGGGUUGUCCGCCUGUCGCUCAUUAGCGGCGCCCCC------UCCGCUGAUGUGGCUGCUCCUGCUGCAGUUGCUUCACCUCCUCCUUCUCCUCCUCCUUCAGUCCAUCAAGUAUUUGUUAUUAAAUAUACGUGUGG ((.((..(((((.....)))))..))))((((((((.(....)------.))))))))(..(((((.......)))))..)..............................(((.((.(((((((....)))))))...))))) ( -36.30, z-score = -2.08, R) >droEre2.scaffold_4784 8398791 129 - 25762168 GACGCCUCUGGCUGUCCGCCUGUCGCUCAUUAGCGGCGCCCCCGAAGUCUCCGCUGAUGUGGCUGCUCCUGCUGCAGUUGCUCCUCCUCCUCC---------------AGUCCGUCGAGUGUUUGUUAUUAAAUAUACGUGUGG (((((..(.(((.....))).)..))..((((((((.(.(......).).))))))))(..(((((.......)))))..)............---------------.)))((.((.(((((((....))))))).))))... ( -35.40, z-score = -0.47, R) >droAna3.scaffold_13337 9163585 115 - 23293914 ---GCCUCGGGA-GUCCGCUUGGCGCUCAUUAGCGGCGCCCCCGAAGACUCUCCUUG-GUGGCAGACUCUCCUUUAGC-AUCACUCCUCCUGCCCC-----------CGGGUUAUUAAGCAUAUACUUGUGG------------ ---((...((((-(((((...((((((.......))))))..))..)))))))((((-(.(((((.............-..........))))).)-----------)))).......))............------------ ( -34.10, z-score = 0.35, R) >consensus GACGCCUCGGGUUGUCCGCCUGUCGCUCAUUAGCGGCGCCCCCGAAGUCUCCGCUGAUGUGGCUGCUCCUGCUGCAGUUGCUCCACCUCCUCCUCC____________AGUCCAUCAAGUAUUUGUUAUUAAAUAUACGUGUGG ...((..(((((.....)))))..))..((((((((..............))))))))((((((((.......)))))))).....................................(((((((....)))))))........ (-21.38 = -22.07 + 0.69)
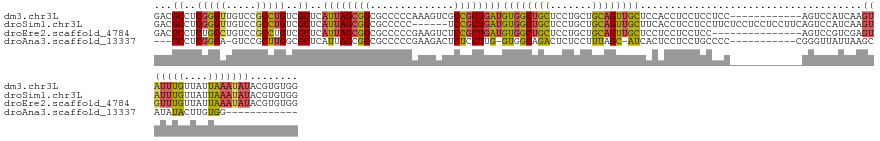

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:18 2011