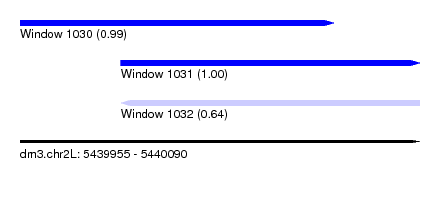
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,439,955 – 5,440,090 |
| Length | 135 |
| Max. P | 0.997539 |

| Location | 5,439,955 – 5,440,061 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.80 |
| Shannon entropy | 0.38350 |
| G+C content | 0.38917 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -17.91 |
| Energy contribution | -17.87 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
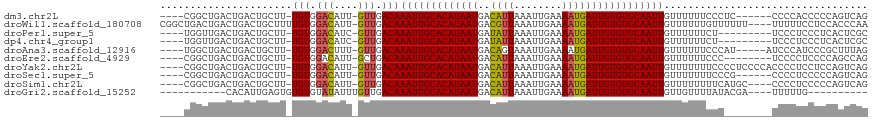
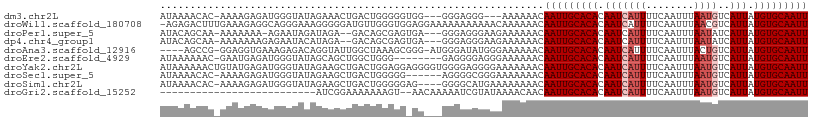
>dm3.chr2L 5439955 106 + 23011544 ----CGGCUGACUGACUGCUU-UGUGGACAUU-GUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCCUC------CCCCACCCCCAGUCAG ----.......(((((((...-.((((.....-...(((((.(((((((((((..((((........)))))))))))))))))))).........------..))))...))))))) ( -30.41, z-score = -3.96, R) >droWil1.scaffold_180708 584442 113 + 12563649 CGGCUGACUGACUGACUGCUUUUGUGGACAUU-GUUGACAAAUUGCACAUAAUGACGUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUGUUUUUU----UUUUUCCUCCACCCAA .((.((.........(..(....)..)(((..-...(((((.(((((((((((..((((........))))))))))))))))))))...))).....----.........)).)).. ( -22.80, z-score = -1.19, R) >droPer1.super_5 6337188 103 - 6813705 ----UGGUUGACUGACUGCUU-UGUGGACAUC-GUUGACAAAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCU---------UCCCUCCCUCACUCGC ----..((..((.((.((((.-...)).))))-))..)).(((((((((((((.((.((......)).)).))))))))))))).........---------................ ( -21.90, z-score = -1.52, R) >dp4.chr4_group1 4741506 103 + 5278887 ----UGGUUGACUGACUGCUU-UGUGGACAUC-GUUGACAAAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCU---------UCCCUCCCUCACUCGC ----..((..((.((.((((.-...)).))))-))..)).(((((((((((((.((.((......)).)).))))))))))))).........---------................ ( -21.90, z-score = -1.52, R) >droAna3.scaffold_12916 6466705 107 - 16180835 ----UGGCUGACUGACUGCUU-UGUGGACUUU-GUUGACAAAUUGCACAUAAUGACAGUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCCAU-----AUCCCAUCCCGCUUUAG ----.......((((..((..-.((((....(-(..(((((.(((((((((((.((((....)))....).))))))))))))))))......)).-----...))))...)).)))) ( -20.50, z-score = -0.50, R) >droEre2.scaffold_4929 5524050 104 + 26641161 ----CGGCUGACUGACUGCUU-UGUGGACAUU-GCUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCC--------UCCCCUCCCCAGCCAG ----.(((((.......((..-((....))..-)).(((((.(((((((((((..((((........)))))))))))))))))))).......--------.........))))).. ( -29.10, z-score = -3.87, R) >droYak2.chr2L 8566904 112 - 22324452 ----CGGCUGACUGACUGCUU-UGUGGACAUU-GUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUCCCCUCCCCACCCCUCCUCCAGUCAG ----...(((((((.......-.((((.....-...(((((.(((((((((((..((((........))))))))))))))))))))............))))........))))))) ( -29.24, z-score = -3.46, R) >droSec1.super_5 3514213 106 + 5866729 ----CGGCUGACUGACUGCUU-UGUGGACAUU-GUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUCCCG------CCCCUCCCCCAGUCAG ----...(((((((.......-.((((.....-...(((((.(((((((((((..((((........))))))))))))))))))))......)))------)........))))))) ( -30.23, z-score = -3.74, R) >droSim1.chr2L 5242743 108 + 22036055 ----CGGCUGACUGACUGCUU-UGUGGACAUU-GUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUUCAUGC----CCCCUCCCCCAGUCAG ----.......(((((((...-.(.((.(((.-(..(((((.(((((((((((..((((........))))))))))))))))))))..)....))).----)).).....))))))) ( -28.80, z-score = -2.82, R) >droGri2.scaffold_15252 5894436 93 - 17193109 -----------CACAUUGAGUGUGUGUAUAUUUGUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUGUUUUAUACGA----UUUUUG---------- -----------(((((...)))))((((((.....((((((.(((((((((((..((((........)))))))))))))))))))))...)))))).----......---------- ( -23.10, z-score = -1.71, R) >consensus ____CGGCUGACUGACUGCUU_UGUGGACAUU_GUUGACAAAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCC________UCCCUCCCCCAGUCAG ......................(((.(((....))).)))(((((((((((((..((((........))))))))))))))))).................................. (-17.91 = -17.87 + -0.04)
| Location | 5,439,989 – 5,440,090 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.54 |
| Shannon entropy | 0.67228 |
| G+C content | 0.34442 |
| Mean single sequence MFE | -15.48 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.54 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
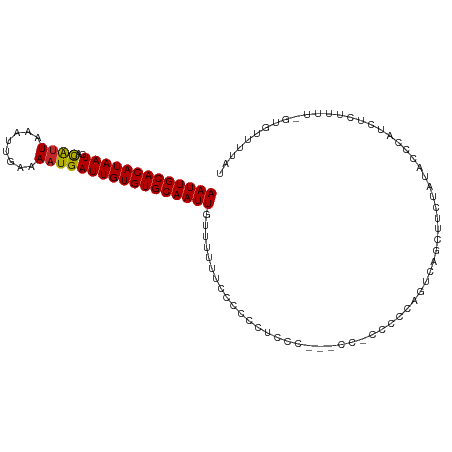
>dm3.chr2L 5439989 101 + 23011544 AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUU---CCCUCCC---CCACCCCCAGUCAGUUUCUAUACCCAUCUCUUUU-GUGUUUUAU (((((((((((((..((((........))))))))))))))))).......---.......---..................................-......... ( -15.10, z-score = -2.32, R) >droWil1.scaffold_180708 584481 107 + 12563649 AAUUGCACAUAAUGACGUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUGUUUUUUUUUUUCCUCCACCCAACAUCCCCCUUUCCCUGCCUCUUUCAAAGUCUCU- (((((((((((((..((((........)))))))))))))))))...............................................................- ( -14.70, z-score = -2.20, R) >droPer1.super_5 6337222 101 - 6813705 AAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCUUCCCUCCC---UCACUCGCUGUC--UCUAUCUAUUCU-UUUUUUU-UUGCUGUAU (((((((((((((.((.((......)).)).))))))))))))).................---............--............-.......-......... ( -15.30, z-score = -3.03, R) >dp4.chr4_group1 4741540 102 + 5278887 AAUUGCACAUAAUGAUAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCUUCCCUCCC---UCACUCGCUGUC--UCUAUGUAUUCUCUUUUUUU-UUGCUGUAU (((((((((((((.((.((......)).)).))))))))))))).................---............--....................-......... ( -15.30, z-score = -2.71, R) >droAna3.scaffold_12916 6466739 102 - 16180835 AAUUGCACAUAAUGACAGUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCCAUAUCCCAU-CCCGCUUUAGCCAAUACCUGUCUCUUUCACCUCC-CGGCU---- (((((((((((((.((((....)))....).)))))))))))))...................-........((((......((.......)).....-.))))---- ( -15.90, z-score = -1.17, R) >droEre2.scaffold_4929 5524084 99 + 26641161 AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCCUCCCCUC--------CCCAGCCAGCUGCUAUACCCAUCUCAUUC-GUUUUUUAU (((((((((((((..((((........))))))))))))))))).................--------..(((....))).................-......... ( -16.80, z-score = -2.79, R) >droYak2.chr2L 8566938 108 - 22324452 AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUCCCCUCCCCACCCCUCCUCCAGUCAGCUUCUAUACCCAUCUCAUACAGUUUUUUAU (((((((((((((..((((........)))))))))))))))))................................................................ ( -15.10, z-score = -2.41, R) >droSec1.super_5 3514247 101 + 5866729 AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUCCCGCCCCU------CCCCCAGUCAGCUUCUAUACCCAUCUCUUUU-GUGUUUUAU (((((((((((((..((((........))))))))))))))))).................------...............................-......... ( -15.10, z-score = -2.12, R) >droSim1.chr2L 5242777 103 + 22036055 AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUUUCAUGCCCC----CUCCCCCAGUCAGCUUCUAUACCCAUCUCUUUU-GUGUUUUAU (((((((((((((..((((........))))))))))))))))).................----.................................-......... ( -15.10, z-score = -1.66, R) >droGri2.scaffold_15252 5894465 80 - 17193109 AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUGUUUUAUACGAUUUUUGUU--ACUUUUUUUCCGAU-------------------------- (((((((((((((..((((........)))))))))))))))))(((((.....))))).......--..............-------------------------- ( -16.40, z-score = -2.28, R) >consensus AAUUGCACAUAAUGACAUUAAAUUGAAAAUGAUUGUGUGCAAUUGUUUUUUCCCCCCUCCC___CC_CCCCCAGUCAGCUUCUAUACCCAUCUCUUUU_GUGUUUUAU (((((((((((((..((((........)))))))))))))))))................................................................ (-14.69 = -14.54 + -0.15)

| Location | 5,439,989 – 5,440,090 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.54 |
| Shannon entropy | 0.67228 |
| G+C content | 0.34442 |
| Mean single sequence MFE | -13.72 |
| Consensus MFE | -9.46 |
| Energy contribution | -9.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | 0.06 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
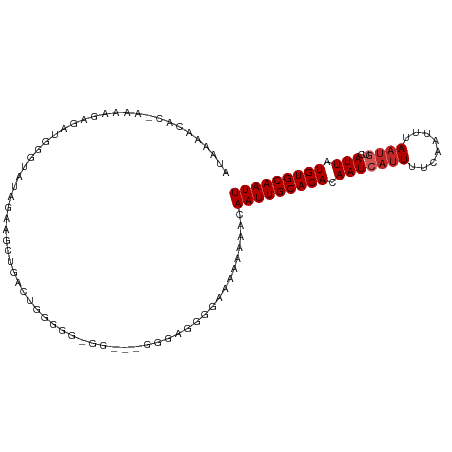
>dm3.chr2L 5439989 101 - 23011544 AUAAAACAC-AAAAGAGAUGGGUAUAGAAACUGACUGGGGGUGG---GGGAGGG---AAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU ......(((-(.((((.((....((.((((.(((.((...(((.---((..(..---......)..)).))).)).))).)))).))...)).)).)).))))..... ( -15.10, z-score = -0.23, R) >droWil1.scaffold_180708 584481 107 - 12563649 -AGAGACUUUGAAAGAGGCAGGGAAAGGGGGAUGUUGGGUGGAGGAAAAAAAAAAACAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAACGUCAUUAUGUGCAAUU -...............(((...(((..(..(((((((.(((.((......................)).))).))).))))..)..)))...)))............. ( -13.95, z-score = 0.50, R) >droPer1.super_5 6337222 101 + 6813705 AUACAGCAA-AAAAAAA-AGAAUAGAUAGA--GACAGCGAGUGA---GGGAGGGAAGAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUAUCAUUAUGUGCAAUU .(((.((..-.......-............--....))..))).---.................(((((((((.(((.((............)).))).))))))))) ( -8.61, z-score = 0.63, R) >dp4.chr4_group1 4741540 102 - 5278887 AUACAGCAA-AAAAAAAGAGAAUACAUAGA--GACAGCGAGUGA---GGGAGGGAAGAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUAUCAUUAUGUGCAAUU .(((.((..-....................--....))..))).---.................(((((((((.(((.((............)).))).))))))))) ( -8.57, z-score = 0.75, R) >droAna3.scaffold_12916 6466739 102 + 16180835 ----AGCCG-GGAGGUGAAAGAGACAGGUAUUGGCUAAAGCGGG-AUGGGAUAUGGGAAAAAACAAUUGCACACAAUCAUUUUCAAUUUACUGUCAUUAUGUGCAAUU ----.(((.-...)))......(((((..((((((....)).(.-.((.(((.((........))))).))..).........))))...)))))............. ( -19.30, z-score = -0.03, R) >droEre2.scaffold_4929 5524084 99 - 26641161 AUAAAAAAC-GAAUGAGAUGGGUAUAGCAGCUGGCUGGG--------GAGGGGAGGGAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU .........-..............((((.....))))..--------.................(((((((((.(((((((........))))..))).))))))))) ( -13.80, z-score = 0.77, R) >droYak2.chr2L 8566938 108 + 22324452 AUAAAAAACUGUAUGAGAUGGGUAUAGAAGCUGACUGGAGGAGGGGUGGGGAGGGGAAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU ........((((((.......))))))...((.(((........))).))..............(((((((((.(((((((........))))..))).))))))))) ( -17.10, z-score = 0.05, R) >droSec1.super_5 3514247 101 - 5866729 AUAAAACAC-AAAAGAGAUGGGUAUAGAAGCUGACUGGGGG------AGGGGCGGGAAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU .........-...................(((..((.....------)).)))...........(((((((((.(((((((........))))..))).))))))))) ( -13.90, z-score = 0.21, R) >droSim1.chr2L 5242777 103 - 22036055 AUAAAACAC-AAAAGAGAUGGGUAUAGAAGCUGACUGGGGGAG----GGGGCAUGAAAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU .........-...................(((..((.....))----..)))............(((((((((.(((((((........))))..))).))))))))) ( -16.00, z-score = -0.41, R) >droGri2.scaffold_15252 5894465 80 + 17193109 --------------------------AUCGGAAAAAAAGU--AACAAAAAUCGUAUAAAACAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU --------------------------..............--......................(((((((((.(((((((........))))..))).))))))))) ( -10.90, z-score = -1.68, R) >consensus AUAAAACAC_AAAAGAGAUGGGUAUAGAAGCUGACUGGGGG_GG___GGGAGGGGAAAAAAAACAAUUGCACACAAUCAUUUUCAAUUUAAUGUCAUUAUGUGCAAUU ................................................................(((((((((.(((((((........))))..))).))))))))) ( -9.46 = -9.86 + 0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:02 2011