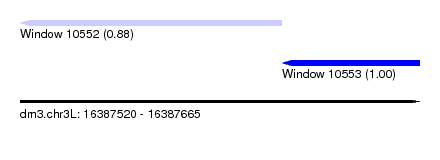
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,387,520 – 16,387,665 |
| Length | 145 |
| Max. P | 0.995539 |

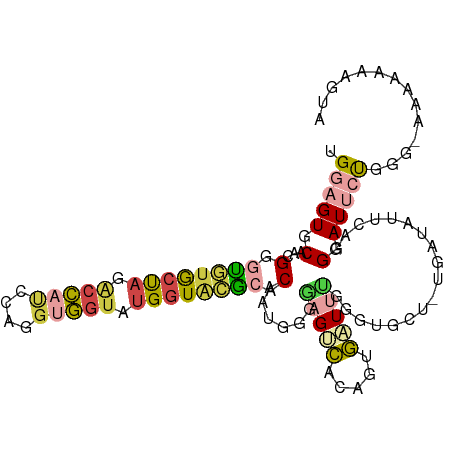
| Location | 16,387,520 – 16,387,615 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 62.24 |
| Shannon entropy | 0.74008 |
| G+C content | 0.49735 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.87 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.56 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
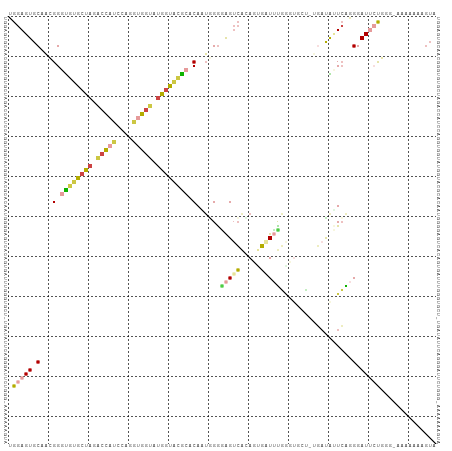
>dm3.chr3L 16387520 95 - 24543557 UGGAGUGCAACGGGUAUACUAGACCAUCCAGGUGGUAUGGUUCGCACAAUGGGUAGUCACAGUGAUAU-----------CAAUCGAGGAUUCUGGG-AAAAUAAGUA .(((((.(....((((((((.(((.(((((.((((......))))....))))).)))..))).))))-----------)......).)))))...-.......... ( -20.70, z-score = 0.33, R) >droSim1.chr3L 15731437 106 - 22553184 UGGAGUGCAACGGGUGUGCUAGACCAUCCAGGUGGUAUGGUACGCACAAUGGGGAGUCAAAGCGAUUUGGUUGCU-GGAUAUUCAGGGAUUCUGGGAAAAAAAAGUA .(((((.(.....((((((((.(((((....))))).))))))))....((((..(((..((((((...))))))-.))).)))).).))))).............. ( -29.70, z-score = -1.52, R) >droSec1.super_0 8481826 105 - 21120651 AGGAGUGCAACGGGUGUGCCAGACCAUCCAGGUGGUAUGGUACGCACAAUGGGGAGUCACAGUGAUUUGGUGGCC-UGAUAUUCAGGGAUUCUGGG-AAAAUAAGUA .....(((.....((((((((.(((((....))))).)))))))).......((((((.(((....)))....((-((.....))))))))))...-.......))) ( -32.70, z-score = -1.04, R) >droYak2.chr3L_random 20677 105 - 4797643 UGGAGUGCUAGGGGUGUACUAGACCACCCAGGGGGUAUGCUACGAUCAGUAGAAAGUCACACUGCUUUGGGUUCU-UGAUGGUCAGGGAUUGUUGG-AAGUAGAGUG ....(((((..(((((........)))))....))))).((((.....)))).....(((.(((((((.((((((-((.....)))))))...).)-)))))).))) ( -31.40, z-score = -0.37, R) >droEre2.scaffold_4784 18633558 106 - 25762168 UGGAGUGCCAGGGGCGUGCUAGGCCACCCAGGUGGUUUGGUACGAACAGUAGGAAGUCGUACUGCUUUGAGUUGUGUGAUGCUCGGGGAUGCUUGC-AAGCAUCGUU ..(((((((((((.(((((((((((((....)))))))))))))..(((((.(....).)))))))))).))........))))...((((((...-.))))))... ( -41.80, z-score = -1.93, R) >droAna3.scaffold_13337 14368996 95 + 23293914 UGGAGUGCGCGGGGUAUGCCAGAUACUAUGUUUUUCCAGGUGUACACAGGUGGGGGGGAAAAGUCUACCAAUAGGAGCAUAUUGGAGGAUAACUA------------ (((.((((.....)))).))).........(((..(((..((....))..)))..)))....((((.((((((......)))))).)))).....------------ ( -22.60, z-score = 0.27, R) >consensus UGGAGUGCAACGGGUGUGCUAGACCAUCCAGGUGGUAUGGUACGCACAAUGGGGAGUCACAGUGAUUUGGGUGCU_UGAUAUUCAGGGAUUCUGGG_AAAAAAAGUA .(((((.(...(.((((((((.(((((....))))).)))))))).)......(((((.....)))))..................).))))).............. (-15.00 = -15.87 + 0.87)

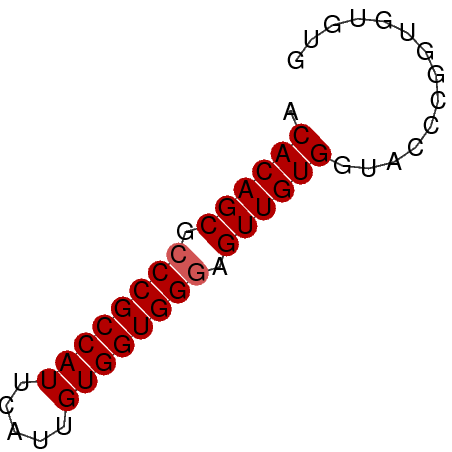
| Location | 16,387,615 – 16,387,665 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 50 |
| Reading direction | reverse |
| Mean pairwise identity | 95.20 |
| Shannon entropy | 0.08663 |
| G+C content | 0.59200 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16387615 50 - 24543557 ACACAGCGCCCGCCAUUCAUUGUGGUGGGAGUUGUGGUACCCGUUGUGUG .((((((.((((((((.....)))))))).)))))).............. ( -23.90, z-score = -3.40, R) >droSim1.chr3L 15731543 50 - 22553184 ACACAGCGCCCGCCAUUCAUUGUGGUGGGAGUUGUGGUACCCGGUGUGUG .((((((.((((((((.....)))))))).)))))).............. ( -23.90, z-score = -2.78, R) >droSec1.super_0 8481931 50 - 21120651 ACACAGCGCCCGCCAUUCAUUGUGGUGGGAGUUGUGGUACCCGGUGUGUG .((((((.((((((((.....)))))))).)))))).............. ( -23.90, z-score = -2.78, R) >droYak2.chr3L_random 20782 50 - 4797643 ACACAGCGCCCGCCAUUCAUUGUGGUGGGAGUUGUGGUACCCGGUGUAUG .((((((.((((((((.....)))))))).))))))((((.....)))). ( -24.60, z-score = -3.34, R) >droAna3.scaffold_13337 14369091 50 + 23293914 ACACAGCGGCCGCCAUUCAUUGUGGUGGGAGUUGUGGUACCUCGUGGGUG .((((((..(((((((.....)))))))..)))))).((((.....)))) ( -22.90, z-score = -2.60, R) >consensus ACACAGCGCCCGCCAUUCAUUGUGGUGGGAGUUGUGGUACCCGGUGUGUG .((((((.((((((((.....)))))))).)))))).............. (-22.80 = -23.00 + 0.20)

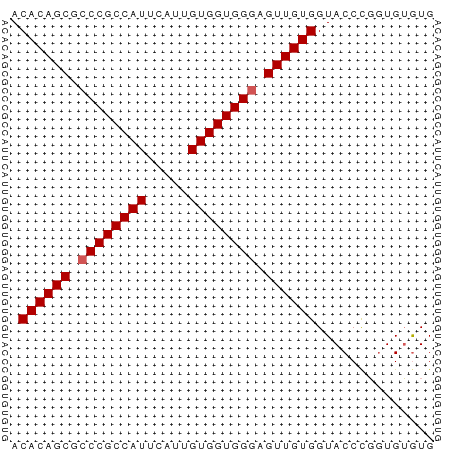
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:12 2011