| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,377,458 – 16,377,554 |
| Length | 96 |
| Max. P | 0.939712 |

| Location | 16,377,458 – 16,377,554 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 62.46 |
| Shannon entropy | 0.67400 |
| G+C content | 0.51120 |
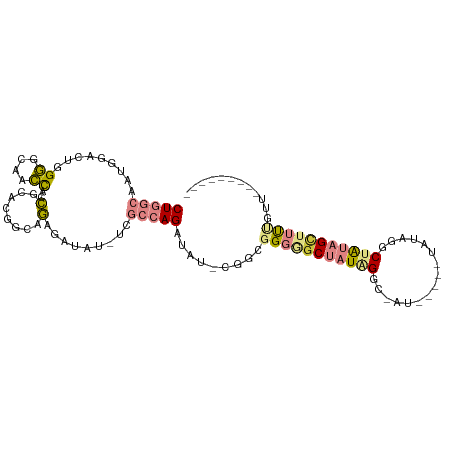
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -12.13 |
| Energy contribution | -13.03 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
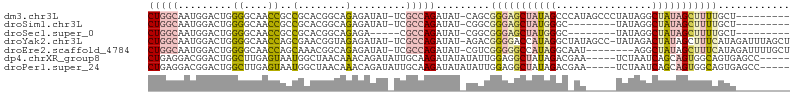
>dm3.chr3L 16377458 96 + 24543557 CUGGCAAUGGACUGGGGCAACCGCCGCACGGCAGAGAUAU-UCGCCAGAUAU-CAGCGGGAGCUAUAGCCCAUAGCCCUAUAGGCUAUAGCUUUUGCU--------- (((((((((..((.((....))(((....)))..)).)))-).)))))....-.((((((((((((((((.(((....))).))))))))))))))))--------- ( -45.00, z-score = -3.88, R) >droSim1.chr3L 15721635 88 + 22553184 CUGGCAAUGGACUGGGGCAACCGCCGCACGGCAGAGAUAU-UCGCCAGAUAU-CGGCGGGAGCUAUGGGC--------UAUAGGCUAUAGCUUUUGCU--------- (((((((((..((.((....))(((....)))..)).)))-).)))))....-.((((((((((((((.(--------....).))))))))))))))--------- ( -39.30, z-score = -3.57, R) >droSec1.super_0 8472143 84 + 21120651 CUGGCAAUGGACUGGGGCAACCGCCGCACGGCAGAGA-----CGCCAGAUAU-CGGCGGGAGCUAUGGGC--------UAUAGGCUAUAGCUUUUGCU--------- (((((.........((....))(((....))).....-----.)))))....-.((((((((((((((.(--------....).))))))))))))))--------- ( -39.10, z-score = -3.81, R) >droYak2.chr3L 2365705 104 - 24197627 CUGGCAAUGGACUGGGGCAACCAGCGAACGGUAGAGAUAU-UCGCCAGAUAU-AGACGGGGACCAUAGGCUAUAGCC-UAUAGACUAUAGCUUUCAUAGAUUUAGCU (((((((((.((((((....))......)))).....)))-).))))).(((-((..(....).((((((....)))-)))...)))))(((...........))). ( -27.20, z-score = -0.75, R) >droEre2.scaffold_4784 18623817 97 + 25762168 CUGGCAAUGGACUGGGGCAACCAGCAAACGGCAGAGAUAU-UCGCCAGAUAU-CGUCGGGGGCCAUAGGCAAU--------AGGCUAUAGCUUUCAUAGAUUUUGCU ..(((((..(((..((....)).......(((..(....)-..)))......-.)))(..(((.((((.(...--------.).)))).)))..).......))))) ( -28.40, z-score = -0.86, R) >dp4.chrXR_group8 2066071 97 + 9212921 CUGAGGACGGACUGGCUUGAGUAAUGGCUAACAAACAGAUAUUGCAAGAUAUAUAUUGGAGGCUAUAGACGAA-----UCUAAUCAGCAGUGGCAGUGAGCC----- ....((.(..((((.((((.((.((((((..(((....(((((....)))))...)))..))))))..))((.-----.....))..))).).))))..)))----- ( -18.80, z-score = 0.43, R) >droPer1.super_24 564275 97 - 1556852 CUGAGGACGGACUGGCUUGAGUAAUGGCUAACAAACAGAUAUUGCAAGAUAUAUAUUGGAGGCUAUAGACGAA-----UCUAAUCAGCAGUGGCAGUGAGCC----- ....((.(..((((.((((.((.((((((..(((....(((((....)))))...)))..))))))..))((.-----.....))..))).).))))..)))----- ( -18.80, z-score = 0.43, R) >consensus CUGGCAAUGGACUGGGGCAACCACCGCACGGCAGAGAUAU_UCGCCAGAUAU_CGGCGGGGGCUAUAGGC_AU_____UAUAGGCUAUAGCUUUUGUU_________ (((((.........((....))...........((......))))))).........(((((((((((................)))))))))))............ (-12.13 = -13.03 + 0.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:11 2011