
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,359,076 – 16,359,194 |
| Length | 118 |
| Max. P | 0.667320 |

| Location | 16,359,076 – 16,359,194 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.03 |
| Shannon entropy | 0.78981 |
| G+C content | 0.55542 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -13.97 |
| Energy contribution | -13.84 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
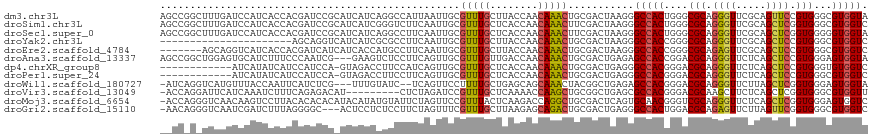
>dm3.chr3L 16359076 118 + 24543557 AGCCGGCUUUGAUCCAUCACCACGAUCCGCAUCAUCAGGCCAUUAAUUGCGUUUGCUUACCAACAAACUGCGACUAAGGGCCACUGGGCGCAGGGUUCGCAGUUCCGUGGGCGUGGUA ..................((((((.(((((.....(((((((.....)).)))))..........((((((((((..(.(((....))).)..)).))))))))..))))))))))). ( -39.90, z-score = -0.51, R) >droSim1.chr3L 15699539 118 + 22553184 AGCCGGCUUUGAUCCAUCACCACGAUCCGCAUCAUCGGGUCUUCAAUUGCGUUUGCUCACCAACAAACUUCGACUAAGGGCCACUGGGCGCAGGGUUCGCAGCUCGGUGGGCGUGGUC ..................(((((((((((......)))))).............(((((((................(.(((....))).).((((.....)))))))))))))))). ( -37.50, z-score = -0.02, R) >droSec1.super_0 8453827 118 + 21120651 AGCCGGCUUUGAUCCAUCACCACGAUCCGCAUCAUCAGGCCUUCAAUUGCGUUUGCUCACCAACAAACUUCGACUAAGGGCCACUGGGCGCAGGGUUCGCAGCUCGGUGGGGGUGGUA ..................(((((..(((.........((((((.(.(((.(((((........)))))..))).).))))))(((((((((.......)).)))))))))).))))). ( -34.70, z-score = 1.06, R) >droYak2.chr3L 2355169 96 - 24197627 ----------------------AGCAGGUCAUCAUCGCGCCUUCAAUUGCGUUUGCUUACCAACAAACUGCGACUAAGGGCCACCGGGCGCAGGGUUCGCAGCUCCGUGGGCGUGGUC ----------------------...........(((((((((......(((((((........)))))(((((((..(.(((....))).)..)).))))))).....))))))))). ( -33.70, z-score = -0.32, R) >droEre2.scaffold_4784 18605779 111 + 25762168 -------AGCAGGUCAUCACCACGAUCAUCAUCACCAUGCCUUCAAUUGCGUUUGCUUACCAACAAACUGCGACUAAGGGCCACCGGGCGCAGAGUUCGCAGCUCCGUGGGCGUGGUC -------....((((........))))......(((((((((......(((((((........)))))(((((((..(.(((....))).)..)).))))))).....))))))))). ( -38.00, z-score = -1.62, R) >droAna3.scaffold_13337 7471101 115 + 23293914 AGCCGGCUGGAGUGCAUCUUUCCCAAUCG---GAAGUCUCCUUCAGUUGCGUUUGUUGACCAACAAACUGCGACUGAGAGCCACCGGACGCAGGGUUCUCAGCUCCGUGGGAGUGGUA ....((((((((.....(((((......)---)))).))))((((((((((((((((....))))))).)))))))))))))(((((((.....))))...(((((...)))))))). ( -44.50, z-score = -1.51, R) >dp4.chrXR_group8 2050529 105 + 9212921 ------------AUCAUAUCAUCCAUCCA-GUAGACCUUCCAUCAGUUGCGUUUGCUCACCAACAAACUGCGACUGAGGGCCACGGGACGCAGGGUUCUCAGCUCCGUGGGUGUGGUC ------------((((((((.....(((.-((.(.(((....(((((((((((((........))))).)))))))))))).)).)))(((.((((.....)))).))))))))))). ( -35.60, z-score = -1.31, R) >droPer1.super_24 548813 105 - 1556852 ------------AUCAUAUCAUCCAUCCA-GUAGACCUUCCUUCAGUUGCGUUUGCUCACCAACAAACUGCGACUGAGGGCCACGGGACGCAGGGUUCUCAGCUCCGUGGGCGUGGUC ------------.................-...((((..((((((((((((((((........))))).)))))))))))(((((((..((.((....)).)))))))))....)))) ( -38.60, z-score = -2.28, R) >droWil1.scaffold_180727 968013 112 - 2741493 -AUCAGGUCAUGUUUACCAAUUCAUCUCG---UUUGUAUC--UCAGUUCCUUUUGCUGAGCAGCAAACUACGGCUGAGAGCCACGGGACGCAGGGUUCUUAGCUCGGUGGGAGUGGUA -.............(((((.....(((((---(((((..(--(((((.......))))))..))))))..((((((((((((.(........))))))))))).))..)))).))))) ( -40.70, z-score = -2.75, R) >droVir3.scaffold_13049 1349987 108 + 25233164 -ACCAGGAUUCAUCAAAUCUUUCAGAGACAU---------CUCUAGAUCCGUUUGCUCAAAACCAAGCUGCGGCUGAGCGCCACGGGACGCAAGCUUCUCAGCUCGGUGGGCGUGGUU -(((((((((.............((((....---------)))).)))))....((((...(((.(((((.((((..(((((...)).))).))))...))))).))))))).)))). ( -33.44, z-score = -0.46, R) >droMoj3.scaffold_6654 83952 117 - 2564135 -ACCAGGGUCAACAAGUCCUUACACACACAUACAUAUGUAUUCUAGUUCCGUUUACUCAAGACCAGGCUGCGACUCAGUGCAACGGGUCGCAGGGUUCUCAGCUCGGUGGGAGUGGUC -....((((.(((..((......))..((((....))))...........))).))))..(((((..(((((((((........)))))))))..((((((......))))))))))) ( -31.60, z-score = -0.11, R) >droGri2.scaffold_15110 2463979 114 + 24565398 -AACAGGGUCAAUCGAUCUUUAGGGGC---ACUCCUCUCCUUCUAGUUUCGUUUGCUUAAGAGCAGACUGCGACUGAGGGCCACUGGACGCAGAGUUCUUAGUUCGGUGGGCGUGGUC -...((((((....)))))).(((((.---...)))))(((((.((((.((((((((....))))))).).)))))))))(((((((((..((....))..)))))))))........ ( -38.60, z-score = -0.60, R) >consensus __CC_G_UUUAAUCCAUCACUACGAUCCG_AUCAUCAGGCCUUCAGUUGCGUUUGCUCACCAACAAACUGCGACUGAGGGCCACGGGACGCAGGGUUCUCAGCUCCGUGGGCGUGGUC ..................................................(((((........)))))...........(((((....(((.((((.....)))).)))...))))). (-13.97 = -13.84 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:09 2011