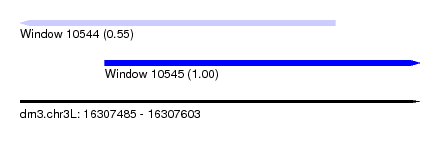
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 16,307,485 – 16,307,603 |
| Length | 118 |
| Max. P | 0.997957 |

| Location | 16,307,485 – 16,307,578 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 61.33 |
| Shannon entropy | 0.78853 |
| G+C content | 0.53710 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -6.59 |
| Energy contribution | -6.28 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16307485 93 - 24543557 UUCAACUGAUGCCACUCGAUGGUGUGGAGGUGGCUUUUAUUCACUUGCCACC-UAGCCAAUUGGCCACCAG--------CAACAGCUGCGGAUUCCGCUGCA---------------- ......((.(((((((....))))((((((((((............))))))-).(((....)))..))))--------)).))((.((((...)))).)).---------------- ( -32.40, z-score = -1.52, R) >droSim1.chr3L 15643544 93 - 22553184 UGCAACUGAUGCCACUCGAUGGUGUGGAGGUGGCUUUUAUUCACUUGCCACC-UAGCCAAUUGGCCACCAG--------CAACAGCUGCGGAUUCCGCUGCA---------------- .(((.(((.(((((((....))))((((((((((............))))))-).(((....)))..))))--------)).)))..((((...))))))).---------------- ( -34.30, z-score = -1.60, R) >droSec1.super_0 8401382 93 - 21120651 UGCAACUGAUGCCACUCGAUGGUGUGUAGGUGGCUUUUAUUCACUUGCCACC-UAGCCAAUUGGCCACCAG--------CAACAGCUGCGGAUUCCGCUGCA---------------- .(((.(((.(((.......(((((..((((((((............))))))-))(((....)))))))))--------)).)))..((((...))))))).---------------- ( -35.41, z-score = -2.22, R) >droYak2.chr3L_random 2743721 93 - 4797643 UACAACUGAUGCCACUCGAUGGUGUGGAGGUGGCUUUUAUUCACUUGCCACC-UAGCCAAUUGGCUACCAG--------CAACAGCUGCGGAUUCCGCUGCA---------------- ......((.(((((((....))))((((((((((............))))))-)((((....)))).))))--------)).))((.((((...)))).)).---------------- ( -33.90, z-score = -2.27, R) >droEre2.scaffold_4784 18550255 93 - 25762168 UGCAACUGAUGCCACUCGAUGGUGUGGAGGUGGCUUUUAUUCACUUGCCACC-UAGCCAAUUGGCCACCAG--------CAACAGCUGCGGAUUCCGCUGCA---------------- .(((.(((.(((((((....))))((((((((((............))))))-).(((....)))..))))--------)).)))..((((...))))))).---------------- ( -34.30, z-score = -1.60, R) >droAna3.scaffold_13337 12972479 72 + 23293914 UGCAACUGAUGCCACUCGACGGCUUGGAGGUGGCUUUUAUUCACUUGCCACC-UCAAACAACAGCUGCCAC--------CA------------------------------------- .(((.(((..(((.......)))...((((((((............))))))-))......))).)))...--------..------------------------------------- ( -25.70, z-score = -3.76, R) >dp4.chrXR_group8 1998886 102 - 9212921 UGCAACUGAUGCCACUCAAUGGUGCAGCGGUGUCUUUUAUUCACUUGCCACCUUACACCGCCGGCUACCUACCCACCGCCAACAGCUGGCAUCUAUGCCACA---------------- .(((...(((((((((...(((((..(((((((.....................))))))).((........))..)))))..)).)))))))..)))....---------------- ( -30.30, z-score = -2.23, R) >droPer1.super_24 497607 102 + 1556852 UGCAACUGAUGCCACUCAAUGGUGCAGCGGUGUCUUUUAUUCACUUGCCACCUUACACCGCCGGCUACCUACCCACCGCCAACAGCUGGCAUCUAUGCCACA---------------- .(((...(((((((((...(((((..(((((((.....................))))))).((........))..)))))..)).)))))))..)))....---------------- ( -30.30, z-score = -2.23, R) >droWil1.scaffold_180698 197836 77 + 11422946 UGCAACUGAUUCCACUUGAGGCUAAUGUGUUGGCUUUUAUUCACUUGCCACU-UAUGCCA----CCAUUUG--------CUGCAGCUGCA---------------------------- .(((.(((....((..((.(((......((.(((............))))).-...))).----.))..))--------...))).))).---------------------------- ( -16.40, z-score = 0.31, R) >droVir3.scaffold_13049 1286698 116 - 25233164 UGCAACUGAUGCCACCGUGACACCACGCGGUGGCUUUUAUACACUUGCCGUCUUGCAUCAGUUGCCAUUGGCUGCAG--CAGCAGCAGCUGCUGCCUCGACGCUCGGCAGUUCCAACA .((((((((((((((((((......)))))))))............((......)))))))))))..((((((((.(--((((((...)))))))..((.....))))))..)))).. ( -50.30, z-score = -3.20, R) >droMoj3.scaffold_6654 27193 114 + 2564135 UGCAACUGAUGCCACCAUGACACCACGCAGUGGCUUUUAUACACUUGGCAUCUUGCAUCAGCUGCCAGUGGCUACAG--CAGUAGCAGCAGCUGCCAC--UGCUCGACAGCCUCAGCA (((((..(((((((..((((..((((...))))...)))).....))))))))))))...((((.....((((..((--((((.((((...)))).))--))))....)))).)))). ( -40.20, z-score = -0.60, R) >droGri2.scaffold_15110 2409094 112 - 24565398 UGCAACUGAUGCCAUCAUAAUACCAUGCAGUGGCUUUUAUACCUUUGUCGUCGUGCAUCAGUUGCCACUGGUUGCUG-CCCCCAGCCGCAGCAGCUGC--CGCUCCACAACAACA--- .(((((((((((.........((.((((((.((........)).))).))).)))))))))))))....((((((((-(........)))))))))..--...............--- ( -35.20, z-score = -1.61, R) >consensus UGCAACUGAUGCCACUCGAUGGUGUGGAGGUGGCUUUUAUUCACUUGCCACC_UACACCAUUGGCCACCAG________CAACAGCUGCGGAUUCCGCUGCA________________ ............................((((((.............................))))))................................................. ( -6.59 = -6.28 + -0.30)

| Location | 16,307,510 – 16,307,603 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 64.81 |
| Shannon entropy | 0.69271 |
| G+C content | 0.53499 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.23 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.60 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.997957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 16307510 93 + 24543557 -----------GGUGGCCAAUUG-GCUAGGUGGCAAGUGAAUAAAAGCCACCUCCACACCAUCGAGUGGCAUCAGUUGAAAGCAGCGAGCUACCAAGACCAA--CCA------------ -----------(((((((....)-)).(((((((............)))))))...))))...(.(((((.((.((((....))))))))))))........--...------------ ( -32.00, z-score = -2.42, R) >droSim1.chr3L 15643569 93 + 22553184 -----------GGUGGCCAAUUG-GCUAGGUGGCAAGUGAAUAAAAGCCACCUCCACACCAUCGAGUGGCAUCAGUUGCAAGCAGCGAGCUACCAAGACUAA--CCA------------ -----------(((((((....)-)).(((((((............)))))))...))))...(.(((((.((.((((....))))))))))))........--...------------ ( -33.10, z-score = -2.37, R) >droSec1.super_0 8401407 93 + 21120651 -----------GGUGGCCAAUUG-GCUAGGUGGCAAGUGAAUAAAAGCCACCUACACACCAUCGAGUGGCAUCAGUUGCAAGCAGCGAGCUACCAAGACCAA--CCA------------ -----------(((((((....)-))((((((((............))))))))..))))...(.(((((.((.((((....))))))))))))........--...------------ ( -34.70, z-score = -3.04, R) >droYak2.chr3L_random 2743746 99 + 4797643 -----------GGUAGCCAAUUG-GCUAGGUGGCAAGUGAAUAAAAGCCACCUCCACACCAUCGAGUGGCAUCAGUUGUAGGCAGCGAGCUACCAAGACCAAGACCAACCA-------- -----------(((.(....(((-(((.((((((............(((((.((.........)))))))....((((....))))..)))))).)).))))...).))).-------- ( -31.80, z-score = -2.20, R) >droEre2.scaffold_4784 18550280 93 + 25762168 -----------GGUGGCCAAUUG-GCUAGGUGGCAAGUGAAUAAAAGCCACCUCCACACCAUCGAGUGGCAUCAGUUGCAAGCAGCAAGCUACCAAGACCAA--GCA------------ -----------(((((((....)-))).((((((............(((((.((.........)))))))....((((....))))..))))))...)))..--...------------ ( -31.80, z-score = -1.86, R) >droAna3.scaffold_13337 12972496 80 - 23293914 -------------------------UGAGGUGGCAAGUGAAUAAAAGCCACCUCCAAGCCGUCGAGUGGCAUCAGUUGCAGGCAGUGAGCCACCGAGACCCA--GUA------------ -------------------------.((((((((............)))))))).......(((.(((((.(((.(((....))))))))))))))......--...------------ ( -30.10, z-score = -3.60, R) >dp4.chrXR_group8 1998911 102 + 9212921 ---GGUGGGUAGGUAGCCGGCGGUGUAAGGUGGCAAGUGAAUAAAAGACACCGCUGCACCAUUGAGUGGCAUCAGUUGCAAGCAGCAAGUCGCCGAGACCCA--CCA------------ ---(((((((.((..((..((((((((.((((................))))..)))))).....(..((....))..).....))..))..))...)))))--)).------------ ( -43.59, z-score = -3.50, R) >droPer1.super_24 497632 102 - 1556852 ---GGUGGGUAGGUAGCCGGCGGUGUAAGGUGGCAAGUGAAUAAAAGACACCGCUGCACCAUUGAGUGGCAUCAGUUGCAAGCAGCAAGUCGCCGAGACCCA--CCA------------ ---(((((((.((..((..((((((((.((((................))))..)))))).....(..((....))..).....))..))..))...)))))--)).------------ ( -43.59, z-score = -3.50, R) >droWil1.scaffold_180698 197855 85 - 11422946 ----------------------GGCAUAAGUGGCAAGUGAAUAAAAGCCAACACAUUAGCCUCAAGUGGAAUCAGUUGCAAGCUGUAAGUCAUCGAACCACCAGCCA------------ ----------------------(((....((((...((((.....(((((((.......((......)).....))))...))).....))))....))))..))).------------ ( -18.60, z-score = -0.56, R) >droVir3.scaffold_13049 1286736 117 + 25233164 --GCUGCAGCCAAUGGCAACUGAUGCAAGACGGCAAGUGUAUAAAAGCCACCGCGUGGUGUCACGGUGGCAUCAGUUGCACGCUGCAAGUCAUCGAGCACCCCACCACUGACUCACACA --..((((((.....(((((((((((......))............(((((((.(......).))))))))))))))))..)))))).......(((((.........)).)))..... ( -41.30, z-score = -0.82, R) >droMoj3.scaffold_6654 27229 108 - 2564135 --GCUGUAGCCACUGGCAGCUGAUGCAAGAUGCCAAGUGUAUAAAAGCCACUGCGUGGUGUCAUGGUGGCAUCAGUUGCACGCUGCAAGCCAUCGAGCACCCCAC-ACAAA-------- --((((((((.....(((((((((((..(((((((((((.........))))...)))))))......)))))))))))..))))).)))...............-.....-------- ( -37.80, z-score = -0.23, R) >droGri2.scaffold_15110 2409126 110 + 24565398 GGGCAGCAACCAGUGGCAACUGAUGCACGACGACAAAGGUAUAAAAGCCACUGCAUGGUAUUAUGAUGGCAUCAGUUGCACGCUGCAAGUCAUCGCGCACCACAC-ACACA-------- ((((.((...((((((((((((((((...........(((......)))....(((.((...)).)))))))))))))).)))))...(....))))).))....-.....-------- ( -35.40, z-score = -1.26, R) >consensus ___________GGUGGCCAAUGG_GCUAGGUGGCAAGUGAAUAAAAGCCACCUCCACACCAUCGAGUGGCAUCAGUUGCAAGCAGCAAGCCACCGAGACCCA__CCA____________ ............................((((((............(((((..............))))).....((((.....))))))))))......................... (-16.89 = -16.23 + -0.66)
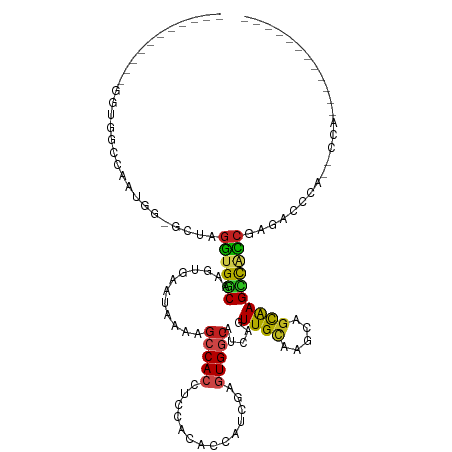
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:31:06 2011